Methods
Sample preparation: Additional method optimization and retention time scheduling was performed using trypsin-digested spike glycoprotein (P0DTC2, SPIKE_SARS2), nucleocapsid protein (P0DTC9, NCAP_SARS2) and QconCAT standard (PolyQuant), prepared as previously described.4
Dilution series were prepared by spiking the recombinant proteins (0, 0.05, 0.1, 0.5, 1, 5, 10, 50, 100 ng) into 250 µL pooled nasopharyngeal swabs from 3 volunteers in eSwab matrix. To all samples, 120 ng QconCAT was added. Samples were vortexed with ice-cold acetone (1750 µL), then centrifuged at 16,000g for 10 mins at 0 °C. Acetone was removed, and pellets were left to dry. Dried pellets were re-suspended in 250 µL of 5% acetonitrile in 50 mM TEAB, then 50 µL of 0.1µg/µL Trypsin/LysC was added. Samples were vortexed, centrifuged and incubated for 15 minutes at 37 °C. 50 µL of each sample was removed and lyophilized. Samples were resuspended in 50 µL of 0.1% formic acid in 5% acetonitrile and 5 µL was injected per analysis. A sample containing approximately 2.5 ng recombinant protein in 250 µL eSwab was prepared by mixing equal volumes of the 5 ng and 0.05 ng samples.
Chromatography: Peptide separation was performed using an ExionLC™ system and a Phenomenex Luna Omega Polar C18 column (2.1 x 50 mm, 1.6 µm). Flow rate of 0.6 mL/min was used for fast analysis times. Gradient is described in Table 1. Column temperature was 40 °C.
Mass spectrometry: MRM analysis of the selected peptides was performed using a SCIEX 7500 System operating in positive ionization mode. Source conditions were ISV 2000 V, GS1 40, GS2 70, TEM 450°C and CUR 40. Scheduled MRM™ Algorithm was used for optimum detection efficiency, using retention time tolerance of 15 sec and a target scan time of 0.4 sec. MRM transitions targeting QconCAT and histone peptides were assigned dwell times of 5 msec. Optimized MRM transitions and compound dependent parameters are shown in Table 2.
Data processing: MRM transition and voltage optimization was performed using Skyline software. Source optimization was performed using SCIEX OS-Q Software. Data were processed using SCIEX OS-Q Software.
MRM assay development
In this work, the MRM assay previously optimized on the SCIEX Triple Quad 5500+ LC-MS/MS System – QTRAP Ready4, targeting peptides from SPIKE and NCAP, was transferred to the SCIEX 7500 System to determine the increased sensitivity. Using the digested recombinant proteins and Skyline software, collision energy values optimized on the SCIEX 5500+ System were re-evaluated on the SCIEX 7500 System and found to be very similar (within 4 V). Stable-isotope labeled standards are routinely used in MS-based assays to increase robustness, allow absolute quantification, and enable normalization across multiple instruments and laboratories. The Cov-MS QconCAT construct includes seventeen 15N-labeled SPIKE and NCAP peptides, separated by their native amino acids and one peptide from each of four human core histone proteins that were added to serve as markers for swab sampling quality. It was surmised that efficient swab sampling should cause superficial tissue damage, resulting in the presence of nuclear histone proteins in the swab sample. If the histone peptides are absent from a sample, this could potentially indicate poor sampling efficiency. MRM transitions were optimized for histone peptides using the QconCAT standard and Skyline software.
The final MRM assay involved two MRMs per peptide (one MRM each for native and heavy-isotope-labeled peptides) for each of ten of the best-responding SPIKE and NCAP peptides and three histone peptides (Table 2). One SPIKE peptide, FLPFQQFGR, found to respond well elsewhere, was included in the assay despite not being present in the QconCAT standard. If this peptide were to be retained for future SARS-CoV-2 antigen analyses, its incorporation into the QconCAT structure would be desirable. Retention times were scheduled, and dwell times fixed at 5 msec for heavy-isotope-labeled and histone peptides, to allow the bulk of the cycle time to be assigned to native SPIKE and NCAP peptides.
Wide-scale detection of SARS-CoV-2 would necessarily require the ability to measure very large numbers of samples per day. In the interest of throughput, the LC column and gradient used for peptide separation was shortened compared to previous work.4 A 50-mm column with 1.6-µm particle size was selected, and the total analysis time shortened to 4.5 min (from 7 min). The shortened analysis time provided good separation of target peptides (Figure 2).
Dilution series
The preservation medium used for the collection of swab samples can have considerable impact on the sensitivity of the LC-MS/MS assay.3 Previously4, samples were analyzed in UTM viral transport medium. However, UTM contains proteins, antibiotics and antimycotics5, which cause high background signal and ion suppression in the assay. ESwab is a salt-based medium without the protein and antibiotic content.6
While ion suppression would still be expected from the eSwab background, no protein-based interference would be anticipated. With this in mind, dilution series were prepared by spiking recombinant proteins and QconCAT into pooled nasopharyngeal swabs from volunteers in eSwab matrix. Samples were subjected to acetone precipitation and protein pellets were resuspended and digested with Trypsin/LysC. Samples were analyzed on the SCIEX 7500 System using the optimized MRM assay.
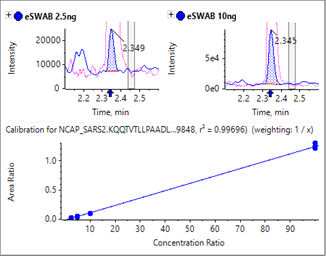
Sensitivity increases of 2 to 20 fold were observed for the SCIEX 7500 System analysis of peptides from SARS-CoV-2 proteins in nasopharyngeal swabs stored in eSwab, when compared with analysis of similar samples in UTM background on the SCIEX 5500+ System4 (Table 3). Lower limits of detection and quantification (LLOD and LLOQ) are outlined in Table 3. Examples of the MRM signal observed from SARS-CoV-2 peptides in eSwab matrix are shown in Figure 3. Calibration curves for all peptides are shown in Figure 4. The typical bioanalytical criteria for determination of LLOQ are accuracy of ±20%, precision of <20% at LLOQ and <15% across the calibration curve.
An example of how use of the QconCAT internal standard can increase assay robustness during sample preparation is shown in Figure 5 and Table 4.
Sensitivity comparison to RNA-based detection
While the work described here is one step further towards a LC-MS/MS assay for the measurement of viral proteins in swab samples, there is still more work to be done. Sensitivities for current RT-PCR tests are measured in cycle threshold (Ct) values. While the same sample can give different Ct values on different RT-PCR assays7 a lower Ct value indicates a higher level of virus. As the amount of virus decreases, Ct values rise gradually. Because Ct is only a semi-quantitative (and variable) indicator of the amount of virus present, the conversion from Ct to amount of viral protein in a swab sample is not straightforward. Nevertheless, recently Van Puyvelde et al.3 estimated a Ct value of 20 to be approximately equivalent to 1.3 fmol NCAP in 10 µL swab sample, and Ct 25 to be equivalent to 41 amol NCAP in 10 µL swab sample. If a 32-fold concentration factor were applied at the sample preparation stage, then 1.3 fmol and 41 amol would correspond to Ct 25 and 30, respectively. These limits of detection in matrix are achievable using high-end triple quadrupole instruments8 but, for the highest possible levels of sensitivity, optimization of sample preparation will be key. Potential areas for improvement include the substitution of eSwab matrix for one with lower salt content, the use of other sample clean-up approaches and the use of sample enrichment strategies. One such strategy, SISCAPA (Stable Isotope Standards and Capture by Anti-Peptide Antibodies), has previously been used to improve the performance of mass spectrometry for measurement of pre-selected protein targets.9
Conclusions
Here, the targeted peptide quantification assay described previously4 has been refined and developed to investigate the potential use of nasopharyngeal swabs in a saline matrix for the quantification of SARS-CoV-2 coronavirus proteins. Further optimization included the addition of a QconCAT stable isotope labeled standard for increased robustness, swab sampling efficiency, inter-laboratory comparison and reduction in analysis time for use in high-throughput environments. The gains in sensitivity attained by moving the assay onto the SCIEX 7500 System, and by reducing matrix effects by changing swab storage medium, could be further increased by use of sample enrichment and clean-up strategies, such as SISCAPA9 or SPE, or by moving to a more MS-friendly swab storage matrix.
This assay has not been validated on real test samples and should not be used for patient testing. More work is required to translate this peptide quantification assay into a useful assay for determination of viral particle numbers.

 Click to enlarge
Click to enlarge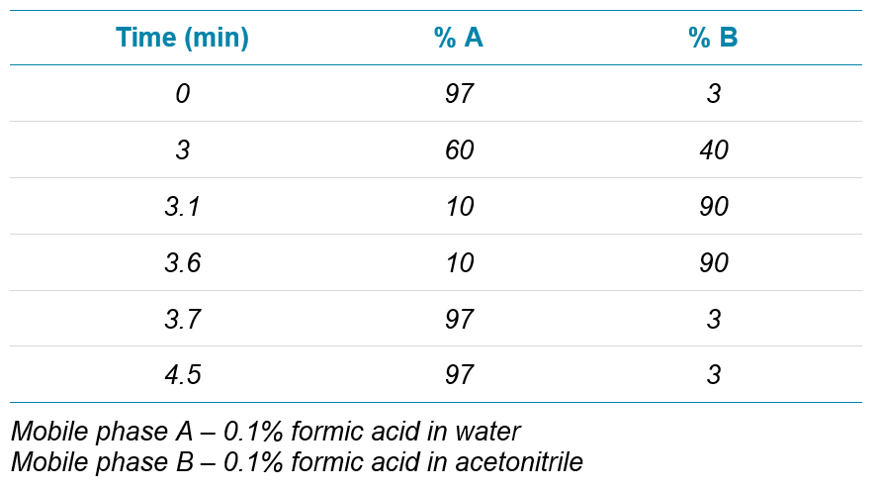 Click to enlarge
Click to enlarge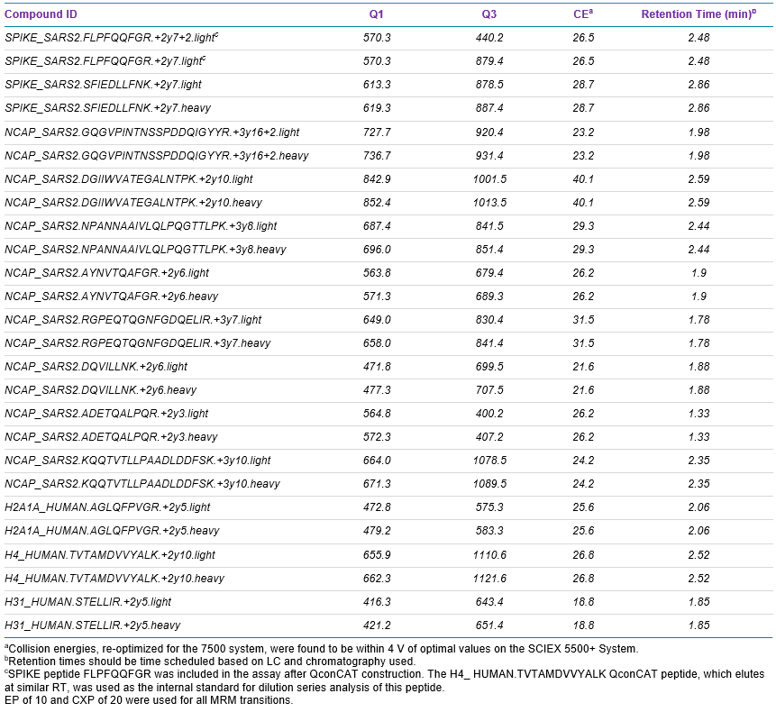 Click to enlarge
Click to enlarge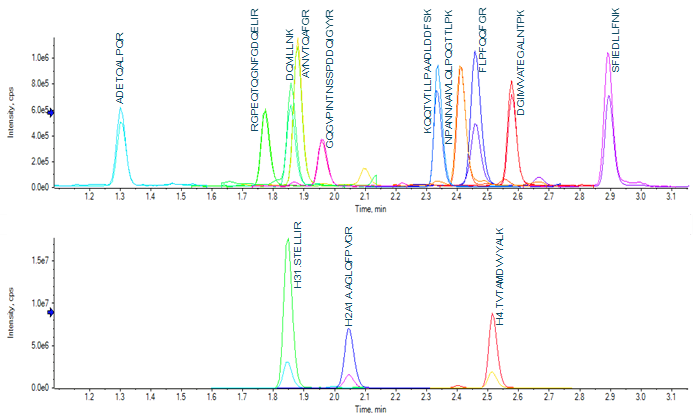 Click to enlarge
Click to enlarge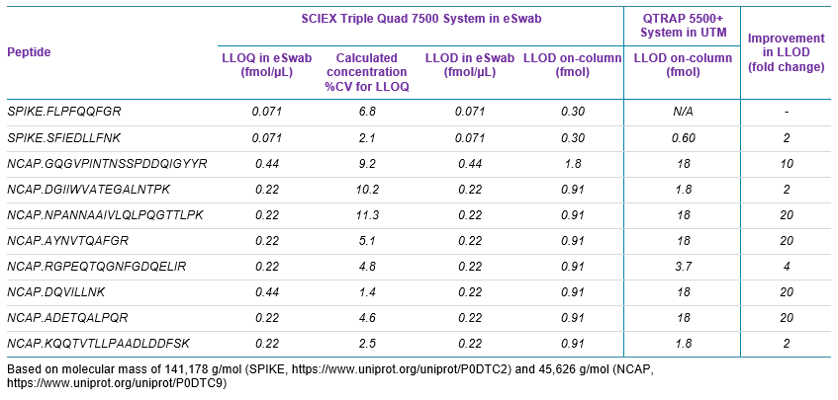 Click to enlarge
Click to enlarge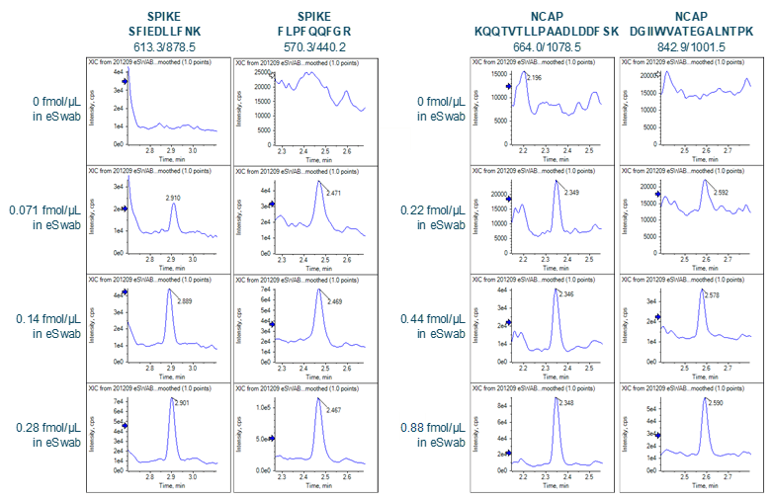 Click to enlarge
Click to enlarge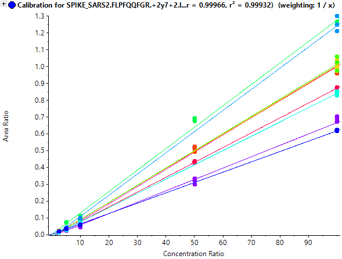 Click to enlarge
Click to enlarge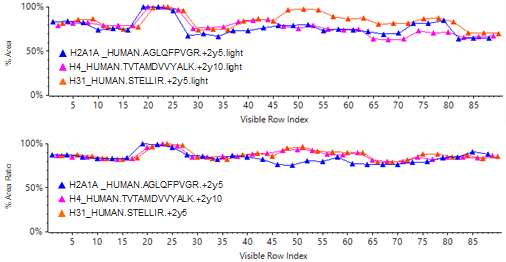 Click to enlarge
Click to enlarge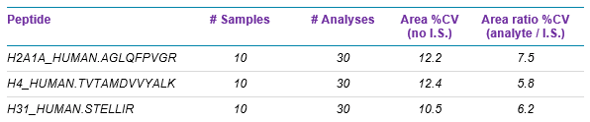 Click to enlarge
Click to enlarge