Factors affecting capillary zone electrophoresis (CZE) separation of charge isoforms of NANOBODY® samples
Abstract
This technical note demonstrates the effect of CZE buffer components on the separations of charge variants from Nanobody samples and evaluates the SCIEX CZE kit to reduce testing variability. This work was developed to help users to implement CZE methods with less development and high reproducibility.
A single NANOBODY domain is approximately 10% of the size of a standard monoclonal antibody.1,2 Multivalent NANOBODY molecules are generated by linking individual NANOBODY domains together.
During production, samples may frequently experience chemical modifications. Processes such as succinimide formation can result in basic isoforms, whereas changes such as deamidation or pyroglutamate formations can lead to acidic charge variants. The assessment of these product quality attributes (PQAs) is important, as modifications can impact the overall charge on the protein, influence protein folding, and potentially reduce the stability and potency of the drug.3
In this technical note, the components of CZE buffers and buffer pH were optimized to achieve the optimal separation following the design of the experiment principle. Assay reproducibility was assessed intra- and inter-batches. SCIEX CZE kit was investigated to achieve similar separation and higher reproducibility compared to in-house prepared buffer.
Key features of CZE separation of NANOBODY samples
Design of experiments (DoE) study:
- Demonstrate CZE buffer preparation optimization to achieve better separation on charge variants
- Investigate the effect of buffer pH on CZE separation of Nanobody samples
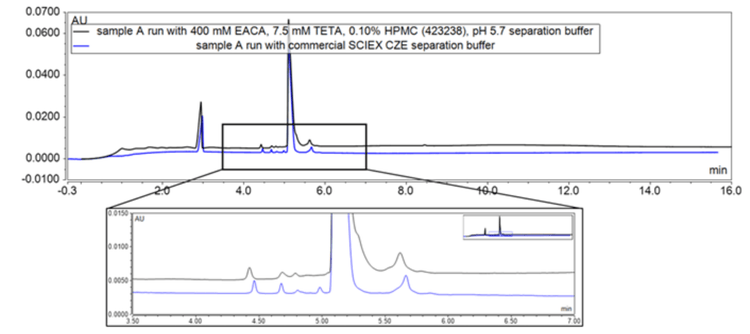
Minimize method development and increase reproducibility: The adoption of the commercially available SCIEX CZE buffer minimizes method development and improve reproducibility for Nanobody sample analysis (see Figure 1)
Introduction
CZE is a widely adopted technique for separating and detecting charge variants in biopharmaceuticals. CZE was originally developed in 2011 by He et al.3 and subsequently subjected to industry ring trials in 2015.4 CZE is a robust method suitable for implementation in a GMP environment. Until recently, CZE separation buffers were unavailable commercially, requiring each laboratory to prepare their buffers, potentially introducing variation into the analysis process.
Methods
Sample preparation: NANOBODY samples (45–75 kDa), originally at 10 mg/mL in formulation buffer containing amino acids (pH 6–7.4), were diluted 10-fold with Milli-Q water before injection. Samples were stored in the autosampler during a batch analysis at 12°C for a maximum of 1 day.
CZE separation method Samples were separated using a PA 800 Plus system (SCIEX). Before samples were tested, a bare fused silica capillary (length 30.2 cm, effective length 20 cm, 50 µm ID, SCIEX P/N: 338451) was conditioned with 0.1M HCl (5 min, 50 psi). Various test buffer solution combinations (5 min, 50 psi) were run, followed by a voltage conditioning step (30 min, 30 kV, normal polarity, 0.17-min ramp)
Samples were then analyzed using a method that initially involved capillary equilibration by rinsing with 0.1M HCl (5 min, 50 psi) and various CZE separation buffer solutions (3 min, 40 psi). All CZE buffer solutions contained mixtures of aminocaproic acid (EACA), triethylenetetramine (TETA), hydroxypropyl cellulose (HPC, ~370 kDa, P/N: 191892) or hydroxypropyl methyl cellulose (HPMC, ~86 kDa, P/N: H7509) from Sigma. The commercially available buffer used was from the CZE Rapid Charge Variant Analysis kit (SCIEX, P/N: C44790). The sample was pressure injected (10 sec, 0.7 psi), followed by a post injection of water (10 sec, 0.1 psi). The sample was separated using normal polarity (16 min, 30 kV, 1-min ramp) at 25°C.
At the end of a batch, a shutdown method was run that involved a rinse with 0.1M HCl (5 min, 50 psi) and water (5 min, 50 psi). An additional water rinse (10 min, 100 psi) was then performed.
Results and discussion
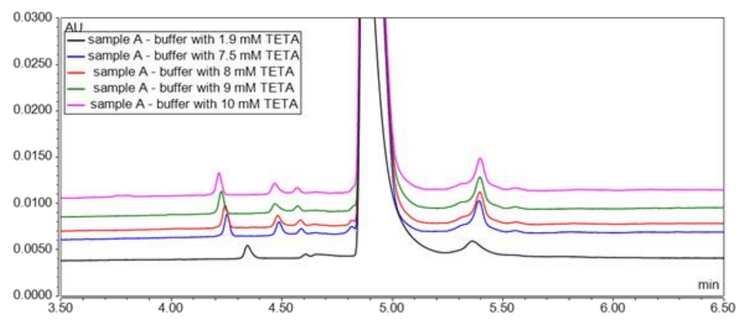
This project studied the effect of CZE buffer components on the separation of NANOBODY samples and their charge variants using the principles of design of experiment (DoE), modifying only 1 constituent at a time. During the study of in-house prepared buffers, the separation method remained the same, employing the EACA consistently at a 400mM, as described in the literature.3 In the first study, the TETA concentration varied from 1.9mM to 15mM with the percentage of HPC set at 0.05% and the CZE buffer at pH 5.7. The pH of the CZE buffer was adjusted by adding 50% acetic acid. As TETA is positively charged, it is used in CZE methods to reduce interaction of analytes with the silanol groups on the capillary wall. TETA can reverse the electric osmotic flow (EOF), leading to broad tailing peaks at low TETA concentrations due to wall interactions. Figure 2 shows that a TETA concentration of 7.5mM was optimal.
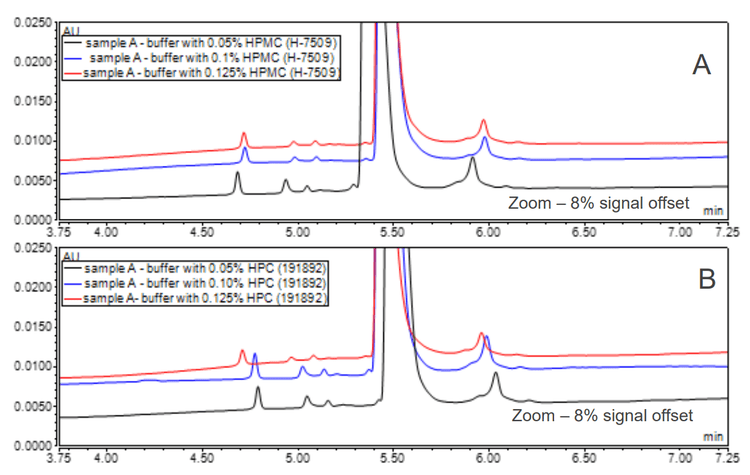
The effect of HPC and HPMC was investigated by injecting different Nanobody samples and varying the size of the polymer used in the buffer, which can impact the buffer viscosity and alter the polymeric sieving. 5 The results indicated that the optimal polymer (HPC or HMPC) varied with the Nanobody sample type. Therefore, it is important to screen the polymer size in method development when developing an in-house buffer.
The amount of polymer used in a buffer is also important. Figure 3 shows a comparison of separation utilizing HPMC and HPC at different concentrations in a CZE buffer. Varying the HPC and HPMC concentrations revealed that 0.1% was the optimal concentration, which was higher than previously reported.3 Combining HPMC and HPC in a buffer did not lead to better separation.
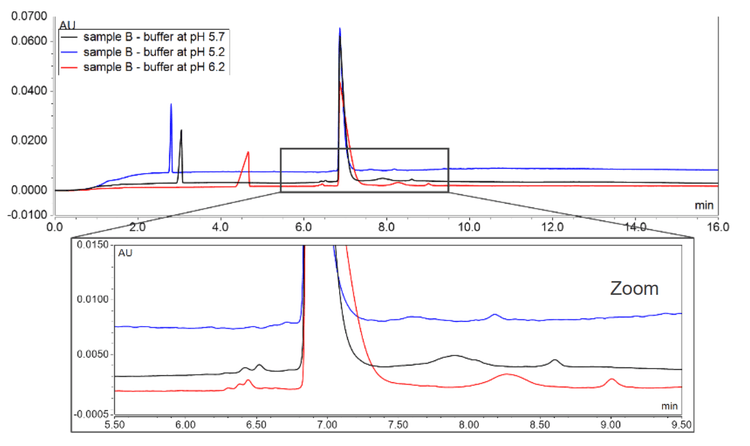
The effect of buffer pH was investigated also. For CZE separation with normal polarity (positive voltage applied at the injection end), the buffer pH should always be lower than the isoelectric point (pI) of the analytes to allow analytes to move in the applied field towards the detector. The NANOBODY samples tested in this project had pI values >7 and therefore, the pH of the buffers used was <7. Figure 4 shows a comparison of 3 buffers with different pH values. As the pH of the buffer approached the pI of the proteins, the peak separations improved. Under these conditions, the peak broadening was also detected, and the later migrating acidic variants were particularly affected. Peak broadening can be reduced by increasing the concentration of TETA, as it acts as a positive dynamic coating reagent and as an ion pairing reagent for acidic species to reduce their wall interactions.
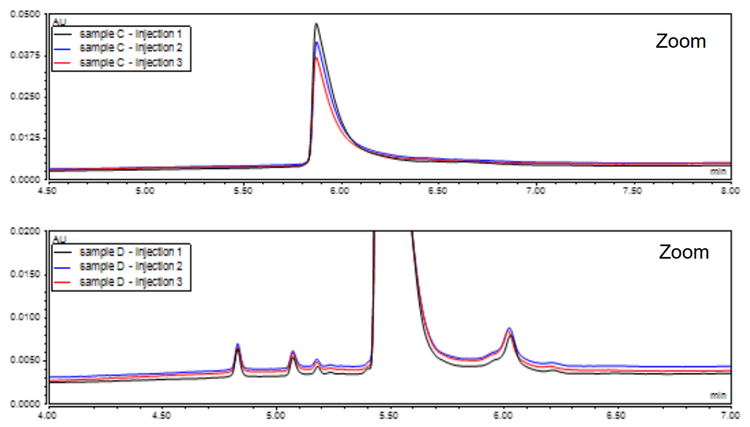
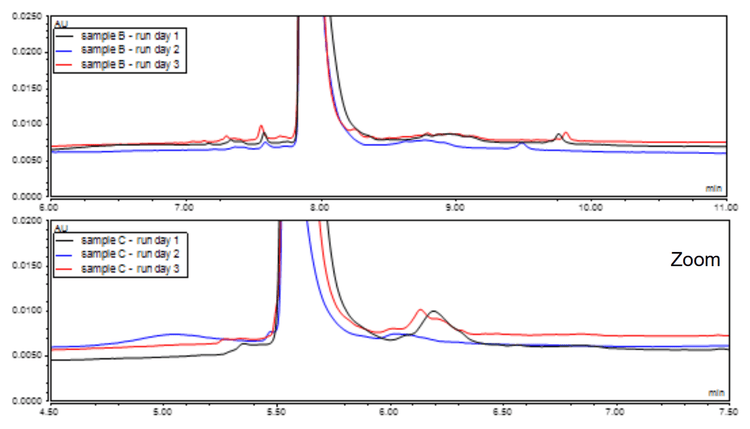
Subsequently, an optimized in-house buffer was assessed for reproducibility within a batch, using a single preparation of the buffer. The observed reproducibility was acceptable, yielding consistent results as depicted in Figure 5. However, some variability was observed when tested across different batches and buffer preparations (Figure 6).
The optimized in-house buffer condition at pH 5.7 was tested compared to the commercial buffer from SCIEX. The key results shown in Figure 1 indicate that the separation obtained for a NANOBODY sample using the commercial kit at a similar pH resulted in an improved peak shape compared to the optimized in-house buffer. Furthermore, more consistent results were observed with the commercial buffer in comparison to the in-house buffer.
Conclusion
- When employing the same in-house CZE buffer preparation, reproducible separation was observed. However, variability was detected between different in-house CZE buffer preparations.
- TETA concentration in an in-house buffer requires optimization to reduce wall interactions and EOF. For NANOBODY samples, the optimal TETA concentration was at 7.5mM.
- The optimal concentration for HPC or HPMC was 0.1% (weight/volume) and different polymers work for different sample types.
- The buffer pH also affects the separation of Nanobody samples. Better separation and broader peaks were observed as the buffer pH approached the pI of the Nanobody molecule.
- The SCIEX CZE Rapid Charge Variant Analysis kit showed better reproducibility and variant separation compared to an optimized in-house buffer
- Use of a SCIEX CZE kit is a good starting point for developing a CZE separation of a NANOBODY sample or another biopharmaceutical
References
- Jovčevska I and Muyldermans S (2020),The Therapeutic Potential of Nanobodies. BioDrugs 34:11–26.
- Ingram JR, Schmidt FI, Ploegh HL (2018), Exploiting Nanobodies’ Singular Traits. Ann Rev Immunol 36:695– 715.
- He, Y., et al. (2011), Rapid analysis of charge variants of monoclonal antibodies with capillary zone electrophoresis in dynamically coated fused-silica capillary, J. Sep. Sci., 34 (5), p.548-555.
- Moritz, B. et al. (2015). Evaluation of capillary zone electrophoresis for charge heterogeneity testing of monoclonal antibodies,J. Chromatogr. B., 983-985, 101- 110.
- Moritz, B. et al. (2017), Optimization of capillary zone electrophoresis for charge heterogeneity testing of biopharmaceuticals using enhanced method development principles. Electrophoresis 38 (24), 3136-3146.