This technical note introduces a novel high-resolution mass spectrometer, the ZenoTOF 8600 system, which was used to identify metabolites in human plasma using an untargeted mass spectrometry approach.
Untargeted metabolomics using a data-dependent acquisition (DDA) scan mode is a common approach to studying the metabolites in biological samples. The experiment is based on a cycle of identifying potential precursor molecules via an MS1 scan and selecting the top candidates for product ion analysis to generate spectra for database matching to identify compounds. To improve the identification of metabolites, two instrument qualities are equally essential: high sensitivity and fast acquisition speed. High sensitivity increases the likelihood of triggering high-quality product ion spectra for low-concentration analytes, and a faster acquisition allows for more DDA cycles during the experiment, improving the chance that the low-level metabolites will be analyzed.
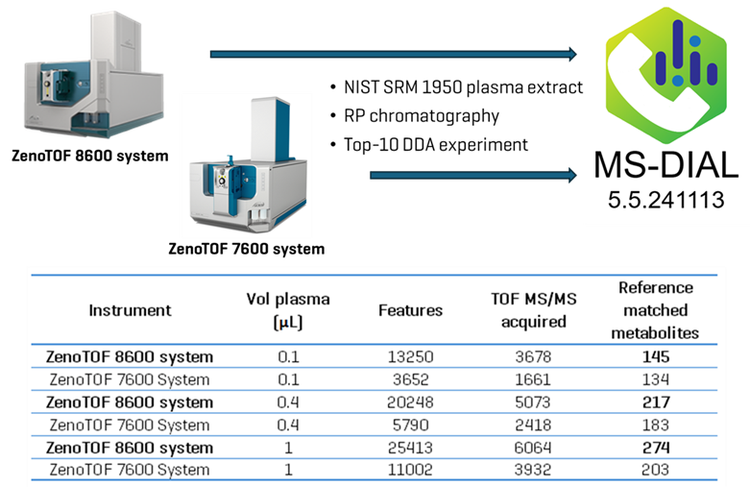
The ZenoTOF 8600 system is a high-resolution QTOF mass spectrometer with a fast analytical speed (~100 MS/MS events/s), and an improved front end to enable a high ion influx paired with a novel optical detector to increase the instrument's sensitivity ~10-fold compared to the ZenoTOF 7600 system, as determined by signal-to-noise (S/N). Here, human plasma was analyzed using the ZenoTOF 7600 and 8600 instruments to evaluate the improved identification of analytes in untargeted metabolomics analysis on human plasma using reversed-phase (RP) chromatography (Figure 1).
Key features of untargeted metabolomics analysis using the ZenoTOF 8600 system
-
Using untargeted analysis, the SCIEX ZenoTOF 8600 system identified 1.4-fold more metabolites than the ZenoTOF 7600 system in human plasma extract (274 vs. 203, analyzing 1 µL plasma equivalents)
-
The ZenoTOF 8600 system demonstrated ~10-fold greater sensitivity at the TOF MS and TOF MS/MS levels than the ZenoTOF 7600 system
Introduction
Metabolomics research has heavily relied on untargeted MS-based analytical methods (i.e., DDA) to detect and potentially quantify small biomolecules within a sample (1,2). High-resolution mass spectrometry (HRMS) analysis using a DDA scan mode is the primary tool for this type of experiment, which is based on an initial survey scan, such as a TOFMS scan, to select potential precursor molecules for product ion analysis. Individual MS/MS scans are acquired for each chosen precursor, and the cycle repeats itself throughout the whole experiment. Algorithms are in place to minimize repetitive measurements of the same compound, but multiple MS/MS spectra for an individual compound are typically acquired. The time for each cycle depends on the number of dependent MS/MS scans, which must be balanced with the chosen chromatographic separation strategy and the speed and sensitivity of the instrument to maximize coverage.
The primary criterion for DDA precursor ion selection is intensity—ions will be chosen in decreasing order from the most intense peak within the survey scan spectrum, provided it has not been excluded based on the experimental exclusion criteria. A plasma-derived metabolomics sample typically has hundreds of precursor ions in each survey scan, so the DDA duty cycle must be as short as possible, the exclusion criteria algorithm carefully balanced, and the instrument must be sensitive for low-level analytes to be present in the spectra for optimal untargeted analysis. The ZenoTOF 8600 system can execute an MS/MS scan in < 10 ms, enabling a scan rate of ~100/s. This rate for a top-10 DDA experiment results in a duty cycle of 600 ms (100 ms for the survey scan and 500 ms for the 50 dependent MS/MS scans). That means for a 6-second-wide eluting peak, there is a strong chance that an MS/MS spectrum would be acquired for any given peak, if it is detected at the precursor ion level. This rate also suggests that for a 20 min gradient, there could be > 40,000 MS spectra acquired. This is not the case. For the ZenoTOF 7600 system, analyzing 0.2 µL plasma equivalents, the number is ~9,000. This means that provided the DDA exclusion criteria were optimally set, the detection capacity of the instrument exceeds the number of detected precursors. It is this aspect of untargeted metabolomics that can benefit from improved instrument sensitivity.
Two instrument qualities are essential for the accurate and comprehensive identification of metabolites using DDA: a high speed of analysis and high sensitivity. Better sensitivity increases the likelihood of triggering product ion analysis for low-concentration analytes, and a faster analysis speed allows for more DDA cycles, which improves the chance that the low-level metabolites will be analyzed. The ZenoTOF 8600 system is a high-resolution QTOF mass spectrometer with fast analytical speed, a reconfigured front end to increase ion flux, and a novel optical detector that combine to increase the instrument's sensitivity ~10-fold compared to the ZenoTOF 7600 system. It is hypothesized that the increased instrument sensitivity will result in an increase in the number of triggered MS/MS events. Furthermore, the MS/MS spectra acquired from the low-level metabolites will be of better quality due to the better sensitivity. Both improvements are expected to increase the number of reference-matched metabolites.
The NIST SRM 1950 human plasma standard was used in these studies to provide a common reference for comparison. Recently, a comprehensive metabolomic/lipidomic profile of the NIST 1950 standard was performed using high-resolution NMR spectroscopy (700 MHz), direct injection tandem mass spectrometry (DI-MS/MS), liquid chromatography-tandem mass spectrometry (LC-MS/MS), and inductively coupled plasma mass spectrometry (ICP-MS) (3). Using these diverse methods, the authors identified and quantified ~ 500 metabolites (lipid molecular species were also quantified, bringing the target list to 1058). The mass spectrometry methods were targeted, which is advantageous in terms of quantitative sensitivity, but they lacked the ability to characterize known- and unknown-unknown compounds. Untargeted analysis is better suited to this type of discovery-based analysis.
A significant challenge associated with untargeted metabolomics is data processing. As is the case for lipidomics data processing, robust interpretation of untargeted metabolomics data is still, effectively, in its early development. For the results presented here, MS-DIAL 5.5 software was used to match DDA-derived MS/MS spectra to its compound database. Because of the diverse programs available to process DDA data and the data themselves having been acquired uniquely despite the sample being the same, direct comparisons of data presented here with literature-based studies should be approached with caution and not necessarily considered an “apples-to-apples” comparison.
Here, human plasma was analyzed using the ZenoTOF 7600 and 8600 instruments to compare the extent and quality of analyte identification in untargeted metabolomics analyses. Due to its improved sensitivity, the ZenoTOF 8600 system identified ~ 1.4-fold more analytes than the ZenoTOF 7600 system under similar analytical conditions and generated superior quality scores, as defined by the MS-DIAL 5.5 software match score.
Materials and methods
Sample preparation: NIST SRM 1950 plasma was extracted using 4 volumes of ice-cold methanol, vortexing for 10 s, centrifuging at 15,000 x g for 10 min, and decanting the supernatant. The supernatant was dried using a speedvac, and the metabolites were resuspended in water to a final concentration of 1 µL extract = 0.1 µL plasma equivalents. The supernatant was directly analyzed by high-performance liquid chromatography electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS).
Chromatography: Samples were analyzed using an Exion LC system with a Kinetex F5 column (2.1 × 150 mm, 2.6 µm, Phenomenex). A simple linear gradient from 0 to 95% B was used with standard reversed phase mobile phases at a flow rate of 200 µL/min. Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. The HPLC rinse solvent was water/methanol/iso-propanol/acetonitrile (1 : 1 : 1 : 1, v/v). The flow rate was 0.200 µL/min, and the gradient conditions are shown in Table 1. A 1 to 10 µL injection volume was used (0.1 to 10 µL plasma equivalents), and the column temperature was maintained at 30°C throughout the analysis. The total runtime was 23 min.
Mass spectrometry: Metabolomics analysis on human plasma extracts was performed on two instruments: a ZenoTOF 7600 system and a ZenoTOF 8600 system, both equipped with an Optiflow Turbo V ion source and an electrospray ionization (ESI) probe. Instrument calibration was maintained using the automated calibrant delivery system (CDS), which calibrated every five samples with an ESI calibration solution specific for the positive ionization mode. DDA experiments were performed using CID-based fragmentation in the positive ion mode.
The systems were configured for CID-based DDA experiments to select the top 10 most abundant ions for fragmentation. Dynamic background subtraction (DBS) with a mass tolerance of 50 mDa was applied to both
experiments to minimize noise and maximize the MS/MS quality. Once a precursor ion was selected and fragmented, it was dynamically excluded from candidate selection for 6 s. The TOF MS accumulation time was set to 100 ms, and the accumulation time for the dependent TOFMS/MS analysis was 50 ms. The TOF MS and TOFMS/MS instrument parameter settings are shown in Table 2. A detailed description of the ZenoTOF 7600 system instrument parameters and their relevance to metabolomics DDA experiments has been previously published (9); the parameter descriptions therein are applicable to the ZenoTOF 8600 system.
To test the ZenoTOF 8600 system's sensitivity improvement, a high-resolution multiple reaction monitoring (MRMHr) scan mode was employed. From the untargeted data, 14 analytes were selected to cover a wide range of masses and peak intensities. The same plasma sample was used, and n=5 replicates were run using the MRMHr scan mode. Instrument parameter settings were the same as those used for the DDA experiments (Table 2).
Data processing: All DDA data were processed using MS-DIAL 5.5 software (4). Optimal parameter settings used for MS-DIAL software processing are presented in a recent SCIEX technical note (5), with some exceptions: the DOT product score and the reverse DOT product score thresholds were set to 500, and the TOFMS threshold was set to 100 counts per second (cps). Metabolite coverage determined from data generated by each instrument is given as reference-matched identifications; the total number of spectra acquired far exceeds these numbers. (For a detailed explanation of match scores, see reference 6.) Quantitative data acquired using the MRMHr scan mode were processed using the Analytics module in SCIEX OS software; Qualitative data were visualized using the Explorer module.
Results
Quantitative performance of the ZenoTOF 8600 system
The ZenoTOF 8600 system was designed with an optical detector that can process more ion current than the detector used in the ZenoTOF 7600 system. This ability, along with the upgraded front-end ion optics, imparts greater sensitivity to the instrument. To measure the impact of improved sensitivity on metabolomics analysis, two different approaches were used: a comparison of the peak areas (and their respective S/N ratios) from a small set of metabolites as measured in the TOFMS dimension, and a targeted MRMHr experiment to measure sensitivity improvements in the MS2 dimension.
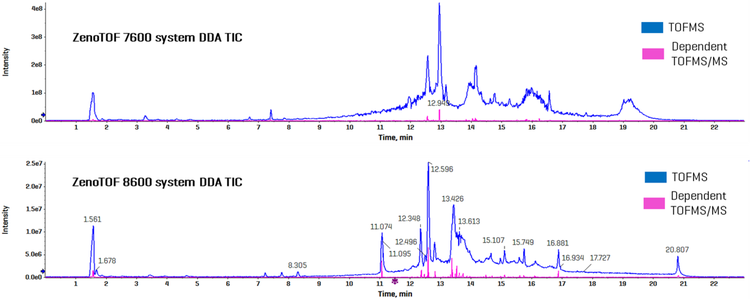
Figure 2 shows the combined TICs (TOFMS in blue and dependent product ion scans in pink) for the ZenoTOF 7600 and 8600 systems (top and bottom panels, respectively). A cursory inspection of each instrument’s TICs indicates an approximately 10-fold increase in ion intensity for the ZenoTOF 8600 system, reflecting the improved sensitivity of the instrument. Because the two instruments perform at the same data acquisition speed, the improved sensitivity is likely responsible for the significant increase in the number of features acquired using the ZenoTOF 8600 system compared to the ZenoTOF 7600 system (25413 vs. 11002 for 1 µL human plasma equivalents; Fig. 1). The improved sensitivity also enabled more triggered dependent scans on the ZenoTOF 8600 system (6064 vs. 3932 MS/MS spectra acquired using 1 µL human plasma equivalents; Fig. 1).
The increased peak intensities acquired on the ZenoTOF 8600 system do not necessarily mean the instrument is more sensitive. Sensitivity improvements should be reported in terms of a relative increase in the S/N. Because DDA experiments are not quantitative at the TOF MS/MS level, sensitivity was assessed by comparing the TOF MS XIC peak intensities and their concordant S/N from metabolites detected on each instrument. An evaluation of the peak areas with S/N calculations for 5 example metabolites is presented in Table 3. The S/N values were calculated using SCIEX OS software using the Explorer data analysis module and represent the average value for each analyte from n=5 injections. The noise region from each TOFMS XIC used to calculate the S/N was selected using the following criteria: the “blank”
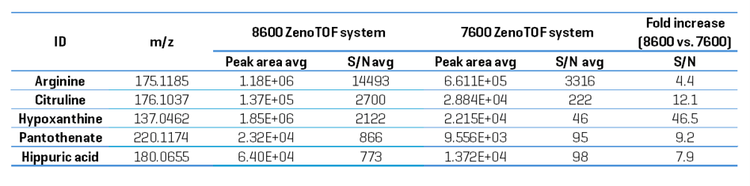
chromatographic region was > 1 min in length and within 2 min of the targeted peak elution time, and the region needed to be devoid of any extraneous isobaric peaks. The S/N of the TOFMS XIC peaks was improved by an average of 16-fold (range = 4.4- to 46-fold) on the ZenoTOF 8600 system compared to the ZenoTOF 7600 system.
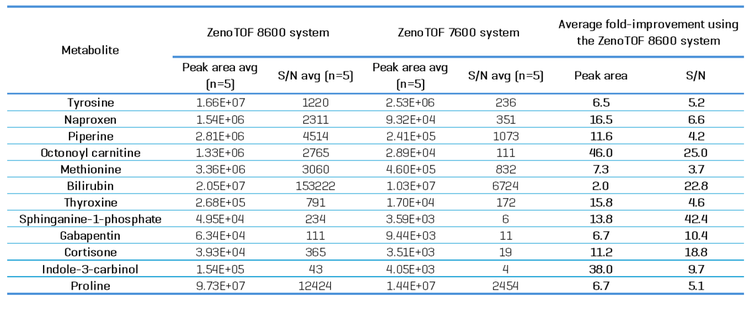
To compare the sensitivity of the two instruments at the MS/MS level, an MRMHr experiment was performed on 12 representative metabolites that cover a broad mass range and diverse peak intensities and areas. Table 4 shows an overall increase in peak areas and calculated s/n values for all compounds, with an average peak area increase of 15.5-fold (range of 2 to 46-fold) and an average S/N value increase of 13.2 (range of 3.7 to 42-fold). These data indicate that the ZenoTOF 8600 system is more sensitive than the ZenoTOF 7600 system for metabolomics studies.
Identification of metabolites using MS-DIAL 5.3 software
The improved sensitivity of the ZenoTOF 8600 system and the increased number of features should translate to better coverage of the metabolome from untargeted metabolomics experiments. In general, better sensitivity allows for detecting less prominent fragment ions, improving the quality of the product ion spectra, and leading to a better match and more confident metabolite identification. To test this hypothesis, DDA experiments were performed on the ZenoTOF 7600 and 8600 systems to compare the numbers of identified metabolites in human plasma lipid extracts.
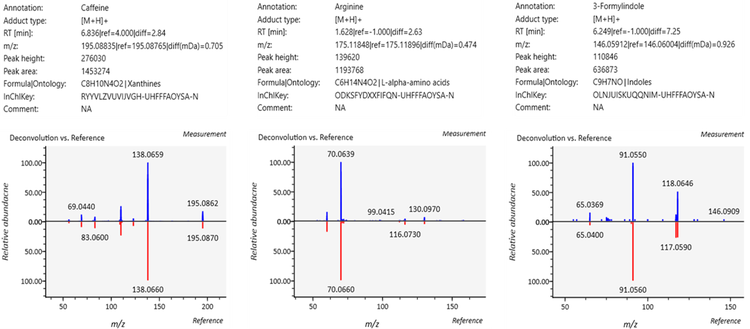
The MS-DIAL user interface provides a comprehensive view of the processed DDA data. Figure 3 shows an abridged example of data that is generated for reference-matched metabolites. Of note is the comparison of experimental data (lower panels, blue spectra) to reference data (red spectra). The enhanced sensitivity of the latter instrument resulted in a 136% increase (~1.4-fold increase) in coverage of the human plasma metabolome in these experiments (274 vs. 203 reference-matched IDs). These identified metabolites have match scores ranging from 2.2 to 1.1. Importantly, in all the comparative experiments performed on the ZenoTOF 7600 and 8600 systems, the latter instrument consistently generated more features, and more MS/MS dependent scans were triggered (Figure 1). This observation is likely due to the enhanced sensitivity in both the MS1 and MS/MS modes of the ZenoTOF 8600 system. The higher MS1 sensitivity logically leads to more candidate ions for the dependent product ion scan, and the enhanced MS/MS sensitivity will impart better fragment intensities and a better overall quality of MS/MS spectrum.
In summary, the ZenoTOF 8600 system is shown to generate superior untargeted metabolomics results compared to the ZenoTOF 7600 system. Although it is challenging to find true equivalent data sets in the metabolomics literature, a study using NIST 1950 plasma analyzed in the positive ion mode, with analytes resolved using RP chromatography, reported 72 distinct metabolites were found using MS-DIAL software for data processing [7], which is less than found here using either instrument. As mentioned above, it is problematic to do a numbers comparison, especially in metabolomics, where there is little standardization and many different software platforms by which the data are analyzed. In the data presented here, the strongest conclusions should be made from the direct comparison of the two instruments’ performances, analyzing the same sample under the same experimental conditions and data processing parameters.
Conclusion
-
In comparative studies, the ZenoTOF 8600 system is ~10-fold more sensitive than the ZenoTOF 7600 system at the TOFMS and TOF MS/MS levels of analysis
-
The SCIEX ZenoTOF 8600 system identified ~1.4-fold more metabolites than the ZenoTOF 7600 system in human plasma extract (275 vs. 203)
-
Using the same analytical method, the improved sensitivity of the ZenoTOF 8600 system generated an increased number of features and dependent product ion scans compared to the ZenoTOF 7600 system (6064 vs. 3932 for 1 µL human plasma equivalents)
References
-
Broeckling, C. D., Beger, R. D., Cheng, L. L., Cumeras, R., Cuthbertson, D. J., Dasari, S., Davis, W. C., Dunn, W. B., Evans, A. M., Fernández-Ochoa, A., Gika, H., Goodacre, R., Goodman, K. D., Gouveia, G. J., Hsu, P. C., Kirwan, J. A., Kodra, D., Kuligowski, J., Lan, R. S., Monge, M. E., … Mosley, J. D. (2023). Current Practices in LC-MS Untargeted Metabolomics: A Scoping Review on the Use of Pooled Quality Control Samples. Analytical chemistry, 95(51), 18645–18654. https://doi.org/10.1021/acs.analchem.3c02924
-
Defossez, E., Bourquin, J., von Reuss, S., Rasmann, S., & Glauser, G. (2023). Eight key rules for successful data-dependent acquisition in mass spectrometry-based metabolomics. Mass spectrometry reviews, 42(1), 131–143. https://doi.org/10.1002/mas.21715
-
Rupasri Mandal, Jiamin Zheng, Lun Zhang, Eponine Oler, Marcia A. LeVatte, Mark Berjanskii, Matthias Lipfert, Jun Han, Christoph H. Borchers, and David S. Wishart. Analytical Chemistry 2025 97 (1), 667-675. https://doi.org/10.1021/acs.analchem.4c05018
-
Takeda, H., Matsuzawa, Y., Takeuchi, M., Takahashi, M., Nishida, K., Harayama, T., Todoroki, Y., Shimizu, K., Sakamoto, N., Oka, T., Maekawa, M., Chung, M. H., Kurizaki, Y., Kiuchi, S., Tokiyoshi, K., Buyantogtokh, B., Kurata, M., Kvasnička, A., Takeda, U., Uchino, H., … Tsugawa, H. (2024). MS-DIAL 5 multimodal mass spectrometry data mining unveils lipidome complexities. Nature communications, 15(1), 9903. https://doi.org/10.1038/s41467-024-54137-w
-
Ozbalci, C., Baker, P. R., & Sayers, R. MS-DIAL software parameters for processing untargeted metabolomics data acquired on the ZenoTOF 7600 system. MKT-30320-A_SCIEX-Technical-Note_metabolomics_ZenoTOF_DDA_parameters-FINAL_1-11-2024.pdf
-
Tsugawa, H., Cajka, T., Kind, T., Ma, Y., Higgins, B., Ikeda, K., Kanazawa, M., VanderGheynst, J., Fiehn, O., & Arita, M. (2015). MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature methods, 12(6), 523–526. https://doi.org/10.1038/nmeth.3393
-
Barbier Saint Hilaire, P., Rousseau, K., Seyer, A., Dechaumet, S., Damont, A., Junot, C., & Fenaille, F. (2020). Comparative Evaluation of Data Dependent and Data Independent Acquisition Workflows Implemented on an Orbitrap Fusion for Untargeted Metabolomics. Metabolites, 10(4), 158. https://doi.org/10.3390/metabo10040158

