Abstract
This technical note presents a quantitative proteomics workflow for low sample volumes using the ZenoTOF 8600 system. Leveraging high sensitivity-driven SWATH data-independent acquisition (DIA), the method enables the identification of >4,900 protein groups and >35,000 precursors from 250 pg of a commercial human digest using nanoflow separations at a throughput of 40 samples-per-day (SPD). The sensitivity of the ZenoTOF 8600 system facilitates robust quantitative biomarker discovery from sub-nanogram sample amounts, making it an ideal platform for single-cell proteomics applications.
Key benefits for quantitative proteomics on low sample volumes using the ZenoTOF 8600 system
- With sensitivity up to 10x greater than previous generation high-resolution instrumentation, the ZenoTOF 8600 system identifies >2x more protein groups and precursors from sub-nanogram sample loadings
- The combination of Whisper Zoom LC methods on the Evosep One system, IonOpticks nanoflow separation technology, and Zeno SWATH DIA on the ZenoTOF 8600 system enables state-of-the-art high-performance nanoflow proteomics at 40 SPD throughput
Introduction
Discovering protein biomarkers often requires analyzing extremely minimal sample loadings, posing a significant challenge for researchers. To overcome this, nanoflow liquid chromatography (LC) combined with SWATH DIA and high-resolution mass spectrometry (HR-MS) enables comprehensive protein profiling, offering both qualitative and quantitative insights from complex biological samples.
The ZenoTOF 8600 system represents a significant leap forward in HR-MS technology, delivering up to a tenfold increase in sensitivity compared to previous-generation instruments. This advancement is driven by the integration of the OptiFlow Pro ion source, enhanced ion capture efficiency, and a next-generation optical detector, which together enable highly sensitive, trace-level detection and robust analysis of complex samples.
In this study, we demonstrate the capabilities of Zeno SWATH DIA on the ZenoTOF 8600 system, paired with Whisper Zoom LC separations on the Evosep One platform. Operating at a throughput of 40 SPD, this setup enables high-sensitivity quantitative proteomics of human protein digests at sub-nanogram levels.
Methods
Sample preparation: Human K562 lysate tryptic digest standard (Promega) was reconstituted in buffer containing 0.1% formic acid/0.05% N-dodecyl β-D-maltoside in water. 250 pg or 500 pg of K562 digest was loaded onto Evotips (Evosep) according to the manufacturer’s instructions.
Chromatography: Separations were performed using the Whisper Zoom 40 SPD method on an Evosep One system. An IonOpticks Aurora Gen3 Elite XS C18 column (15 cm x 75 µm), heated to 60°C, was used for all injections. All experiments were performed in triplicate.
Mass spectrometry: SWATH DIA was performed on a ZenoTOF 8600 system using an ion source configured with a 1-50 µL/min microflow probe. Ion source parameters were as follows: gas 1 was set to 10 psi, curtain gas set to 35 psi, ionspray voltage set to 2000 V, and interface heater temperature set to 300°C. The Zeno SWATH DIA method consisted of a TOF-MS scan from 400 to 1500 Da with an accumulation time of 100 msec, and 85 variable-width Zeno SWATH DIA windows with MS/MS scans from 140 to 1750 Da, each with an accumulation time of 18 msec.
Data processing: Data files were converted to .dia format using a research-grade data converter. Converted files were processed with DIA-NN version 1.9.1 software (1), using a K562/HeLa spectral library described previously (2). Default parameter settings were used with the following exceptions: precursor m/z range was set to 400-900, fragment m/z range was set to 140-1750, and Match Between Runs (MBR) was used. Triplicate files for a given loading were searched together. The number of protein groups and precursors identified and quantified with coefficients of variation (CV) values <20% or <10% were determined from the output “pg_matrix.tsv” and “pr_matrix.tsv” files generated by DIA-NN software.
Results
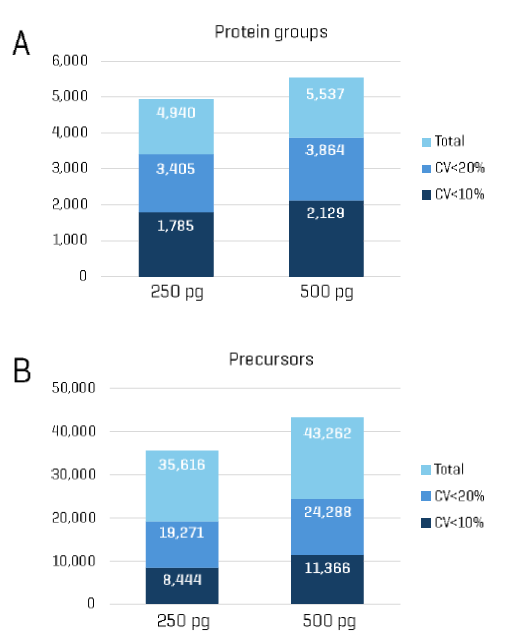
Figure 1 summarizes the identification and quantitation of protein groups and precursors from K562 digest samples at two concentrations: 250 pg and 500 pg. From triplicate injections of the 250 pg sample, >4,900 protein groups and >35,000 precursors were identified. Of these, approximately 70% of protein groups (>3,400) and 54% of precursors (>19,000) were quantified with CVs <20%. Increasing the sample amount to 500 pg improved detection, yielding >5,500 protein groups and >43,000 precursors. Similarly, 70% of protein groups and 56% of precursors were quantified with CVs <20%. It should be noted that protein and peptide identifications from a given loading of bulk commercial digest may differ slightly from the number of identifications achievable from equivalent amounts of single-cell preparations.
Conclusions
- Using Whisper Zoom 40 SPD on the Evosep One system and Zeno SWATH DIA on the ZenoTOF 8600 system, more than 4,900 and 5,550 protein groups were identified from 250 pg and 500 pg K562 digest loadings, respectively.
- These same loadings yielded over 35,000 and 43,000 precursors, respectively.
- Of the identified protein groups and precursors, 70% and 54–56% were quantified and with CVs below 20%, respectively, demonstrating robust quantification even at sub-nanogram levels.
References
1. Demichev et al., Nature Methods, 2020, https://www.nature.com/articles/s41592-019-0638-x.
2. Large-scale protein identification using microflow chromatography on the ZenoTOF 7600 system. SCIEX technical note, RUO-MKT-02-14415-A.