Abstract
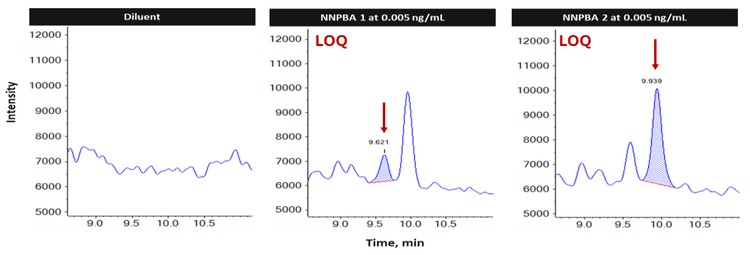
This technical note demonstrates a sensitive method for the quantitation of N-nitroso Pyribenzamine (NNPBA) isomeric impurities (NNPBA 1 and NNPBA 2) in Tripelennamine hydrochloride (TH) API using the QTRAP version of the SCIEX 7500+ system. A limit of quantitation (LOQ) of 0.005 ng/mL was achieved with no interference in the diluent blank and unspiked API samples for NNPBA isomers in a single method (Figure 1).
Tripelennamine hydrochloride (TH) is an ethylenediamine derivative. It is a sedating histamine H1 receptor antagonist with antimuscarinic properties. TH is used to alleviate symptoms of asthma, hay fever and both perennial and seasonal allergic rhinitis, as well as to manage other allergic conditions, such as urticaria.1 TH salt is formulated as tablets and called Pyribenzamine.2,3 It works by blocking the histamine Type 1 (H1) receptor, thereby reducing inflammatory reactions triggered by histamines.4 Regulatory bodies have amended the maximum permissible daily intake limit for NNPBA to 18 ng/day.5 This is equivalent to a maximum daily dose of less than 600 mg/day.1 Given that TH has a maximum daily dose of 600 mg/day, a nitrosamine limit of 0.03 ng/mg is required to be quantified in the API. This technical note presents a reliable and sensitive workflow to support the quantitative analysis of NNPBA in TH API below the regulation limit using the QTRAP 7500+ system.
Key benefits for analysis of NNPBA using the QTRAP 7500+ system
- Sub-ng/mL level of quantitation: Achieve 0.005 ng/mL LOQ for the quantitation of both NNPBA isomeric impurities (NNPBA 1 and NNPBA 2)
- Quantitative sensitivity below the specification limit: NNPBA 1 and NNPBA 2 were analyzed at 0.01 ng/mg of API, below the calculated specification limit of 0.03 ng/mg
- Baseline chromatographic separation: Achieve accurate quantitation with good baseline separation of NNPBA and TH API, as well as accurate quantitation and separation of NNPBA 1 and NNPBA 2 isomers
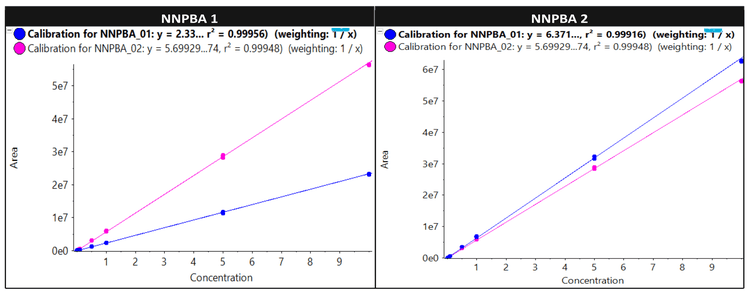
- Robust analytical performance: Accurate quantitative performance with %CV <5 for quantifier ion and qualifier ion was achieved at all concentration levels across a linear dynamic range (LDR) of 3.3 orders of magnitude for NNPBA 1 and NNPBA 2
- Streamlined data management: SCIEX OS software, a 21 CFR Part 11-compliant platform, simplifies data acquisition and processing
Introduction
Nitrosamines are highly potent carcinogens that affect different organs6 and are classified (class 1-5) using the Carcinogenic Potency Categorization Approach (CPCA).7 Here, the severity was determined based on the acceptable intake, activating, or deactivating features defined in the structures. Based on the structure of NNPBA, it is placed under a class 1 category following the CPCA.
Tripelennamine is a tertiary amine class of antihistamines3, causing the formation of NNPBA in TH to be higher. Since the probability of forming nitrosamine impurity is higher, the EU has set a regulation limit of 18 ng/day. Considering the maximum daily dosage of 600.0 mg1, and the regulation limit, NNPBA should be determined below 0.03 ng/mg.
Methods
Standard preparation: Calibration curve dilutions of NNPBA were prepared across a range of concentrations in 0.1% (v/v) formic acid in water (0.005, 0.01, 0.025, 0.05, 0.1, 0.5, 1, 5, and 10 ng/mL) and analyzed in triplicate.
Sample preparation: 1 mg of TH was weighed into a suitable vessel. 1 mL of 0.1% (v/v) formic acid in water was added and vortexed thoroughly to obtain a 1 mg/mL concentration. Quality control samples were prepared by spiking samples of NNPBA in TH API at 3 concentration levels: 0.01 ng/mg, 0.02 ng/mg and 0.03 ng/mg. Six replicates of each concentration were analyzed.
Chromatography: Analytical separation of the isomers, and between the compound and the API, was performed on the ExionLC system using a Phenomenex Kinetex Biphenyl (3.0 × 100 mm, 2.6 μm) column at a flow rate of 0.4 mL/min. Mobile phase A was 0.1% (v/v) acetic acid in water and mobile phase B was 0.1% (v/v) acetic acid in methanol. The column temperature was set to 30°C. The gradient conditions used are summarized in Table 1. An 8 μL sample aliquot was injected for LC-MS/MS analysis. The LC flow was diverted to waste for the first 8 min to prevent TH API from entering the mass spectrometer.
Quantitative performance on the QTRAP 7500+ system
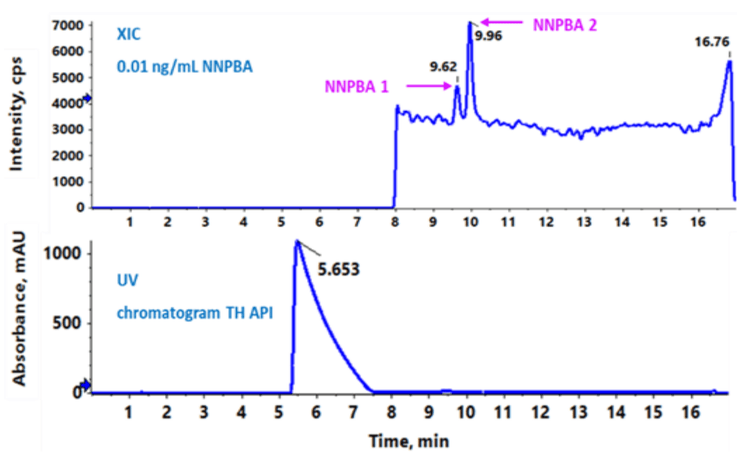
The baseline separation of approximately 3.9 minutes and 4.3 minutes was achieved between the TH and NNPBA 1 peaks, and TH and NPBA 2 peaks, respectively. Based on the sequence of elution, they are addressed as NNPBA 1 and NNPBA 2. The TH API eluted at 5.65 min, while the isomer NNPBA 1 and NNPBA 2 was eluted at 9.62 and 9.96 min, respectively (Figure 2).
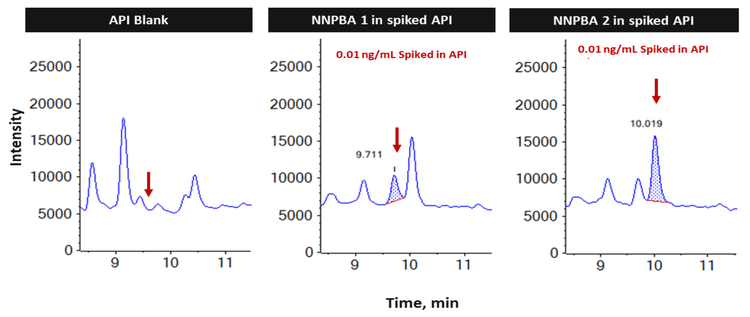
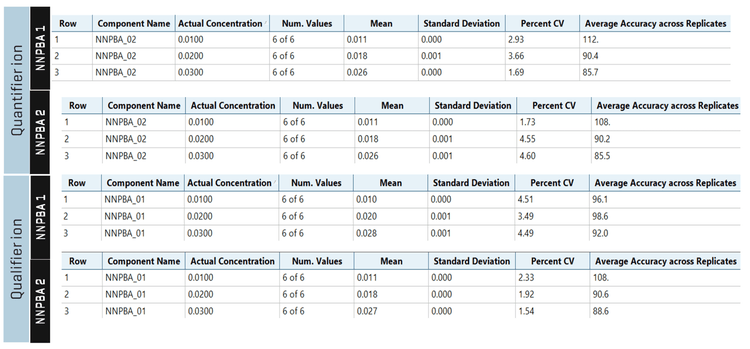
The average recovery of NNPBA 1 and NNPBA 2 was 102.2% and 113.4%, respectively. The %CV of NNPBA 1 and NNPBA 2 was <5% for spiked samples, evaluated in triplicate for neat standard and 6 replicates for the spiked sample (Table 4).
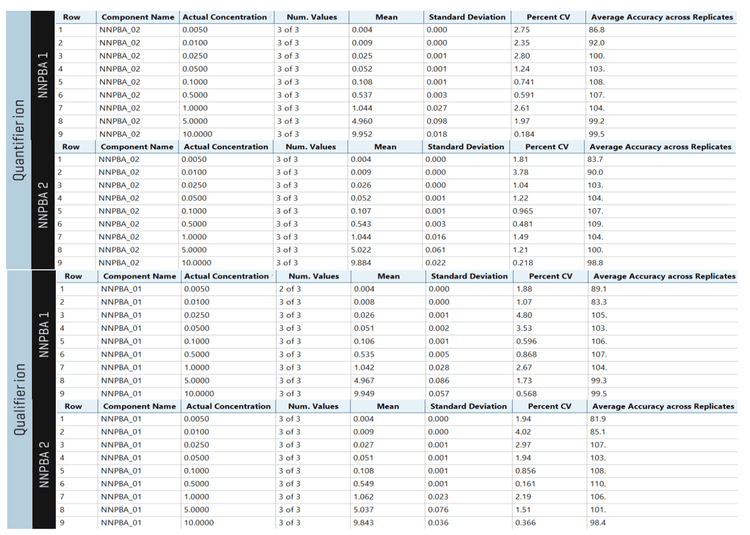
Analytical performance was evaluated based on the criteria that the accuracy of the calculated mean should be between 80% and 120% at the LOQ and between 85% and 115% at the higher concentrations. In addition, the %CV of the calculated mean of the concentration should be <20% at the LOQ and <15% at all higher concentrations.
The assay accuracy for both the isomers was within ±18% at LOQ and ±10% at higher levels of the actual concentration and the %CV was <5 throughout the batch. Calculated percent accuracy and %CV values were within the acceptance criteria at each concentration level for both NPBA 1 and NPBA 2 (Figure 5).
In the spiked quality control samples, all the parameters met the established acceptance criteria, reinforcing the method's reproducibility (Figure 6).
Compliance-ready SCIEX OS software
Equivalent SCIEX OS software capabilities for regulated bioanalysis can be executed on the QTRAP 7500+ system, ensuring high fidelity when performing method transfers while retaining critical compliance features.
SCIEX OS software is a closed system and requires records and signatures to be stored electronically, meeting the regulations outlined by 21 CFR Part 11. SCIEX OS software can open raw data files from any visible storage location within a closed network by using designated processing workstations. Figure 7 illustrates the features of SCIEX OS software used to monitor the audit trail, acquire and process data, and configure user access. The audit trail feature enables users to audit critical user actions and locks in data integrity. The Central Administrator Console (CAC) feature allows users to centralize acquisition and processing using a single platform to maximize efficiency for multi-instrument laboratories, independent of compliance standards. The configuration module allows users to assign roles and access as the administrator, method developer, analyst, and reviewer.

Conclusions
- An LOQ of 0.005 ng/mL was achieved for the quantitation of NNPBA 1 and NNPBA 2
- Linearity was achieved at concentrations ranging from 0.005 ng/mL to 10 ng/mL with an r2 >0.999 for both quantifier and qualifier ions covering LDR of 3.3 orders of magnitude for NNPBA 1 and NNPBA 2
- Good quantitative performance was demonstrated with accurate and highly reproducible (%CV <5%) results on the QTRAP 7500+ system
- An average recovery of >85% was achieved with %CV <5 where NNPBA 1 and NNPBA 2 were analyzed at 0.01 ng/mg of API, below the calculated specification limit of 0.03 ng/mg
- The method demonstrated excellent quantitative performance in the spiked quality control samples for NNPBA 1 and NNPBA 2 (0.01 ng/mg, 0.02 ng/mg and 0.03 ng/mg) using a single method
- A single platform for streamlined data acquisition, processing, and management with SCIEX OS software was presented
- Retain data management and compliance-readiness (21 CFR Part 11) features using SCIEX OS software to support nitrosamine analysis on the QTRAP 7500+ system
References
- Tripelennamine. Kelley K. Kiningham
- Website: www.jamanetwork.com/journals/jama/article-abstract/292869
- Halide counterions in FDA-approved pharmaceutical salts. Chandani T. Muleva, Sonali S. Bharate, in Journal of Drug Delivery Science and Technology, 2023
- Tripelennamine Citrate and Tripelennamine Hydrochloride. Mark G. Papich DVM, MS, DACVCP
- Nitrosamine impurities in medications: Established acceptable intake limits. Appendix 1: Established acceptable intake (AI) limits for N-nitrosamine impurities (version: 2024-05-31).
- Nitrosamines Heriberto Robles
- Carcinogenic Potency Categorisation Approach for Nnitrosamine European Medicines Agency Appendix 2


