Abstract
This technical note describes how the lower limits of quantification (LLOQs) for small molecule pharmaceutical compounds in rat plasma are improved using a high-end triple quadrupole mass spectrometer. Sub-pg/mL quantification was achieved for midazolam, imipramine and clozapine with outstanding reproducibility, precision, accuracy, and linearity. The improved front-end technology of the SCIEX 7500 system enhanced assay sensitivity to meet the demands of routine bioanalysis in complex matrices.
Demands for improved sensitivity in routine bioanalytical assays continue to increase as drug discovery and development programs focus on more efficacious, lower dosage compounds. Sample preparation and alternative chromatography techniques (such as microflow) are often used to meet these assay sensitivity needs, but they can be time consuming. In many cases, use of a more sensitive mass spectrometer is the easiest way to meet these needs, and in combination with the aforementioned techniques, can provide even more assay sensitivity. An increase in the signal generated by a specific concentration of an analyte does not necessarily correspond to an improvement in assay LLOQ. Assay LLOQs are generally limited by signal to noise ratios and the reproducibility of an analyte’s response as it approaches the baseline response. Here, three small molecule pharmaceutical compounds, imipramine, midazolam, and clozapine were quantified in rat plasma. Samples were prepared using simple protein precipitation method and LLOQ (lower limits of quantitation) of 0.050pg/mL for midazolam, 0.100pg/mL for imipramine and 0.200pg/mL for clozapine were achieved using the SCIEX 7500 system.
Introduction
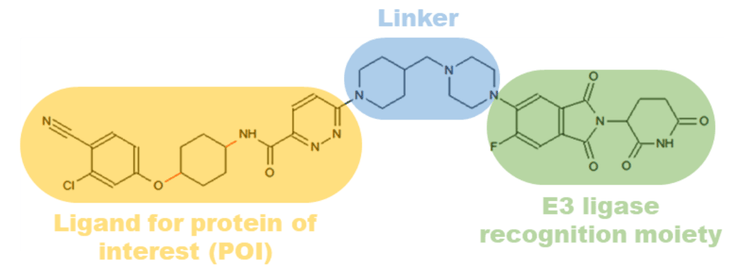
The concept for employing PROTACs for targeted protein degradation was first demonstrated in 2001 by Sakamoto et al.1 Since then, considerable developments have been made to apply this approach to treat drug-resistant targets. 2 PROTACs are heterobifunctional compounds composed of a ligand for a protein of interest (POI) and an E3 ligase that are connected through a linker (Figure 1). PROTACs initiate degradation by forming a ternary complex with the POI followed by ubiquitination. The ubiquitinated POI is then detected and degraded by endogenous 26S proteasomes present in eukaryotic cells.2 The unique chemical knockdown strategy of PROTACs makes it a remarkable technology for targeted protein degradation. Additionally, PROTACs reach complete efficacy at low dosages compared to conventional small molecules. Therefore, highly sensitive bioanalytical techniques are needed to perform pharmacokinetic and pharmacodynamic studies to evaluate product safety and efficacy.
Here, ARV-110 was employed as a model PROTAC to develop a quantification method in rat plasma using the SCIEX 7500 system. The improvement in the front-end technology of the SCIEX 7500 system enabled a lower limit of quantification (LLOQ) of 5 pg/mL to be achieved for ARV-110 using a protein precipitation sample preparation. The assay covered a calibration range covering 5 pg/mL to 5000 pg/mL with a linear dynamic range (LDR) spanning 3 orders of magnitude.
Key features of bioanalysis of PROTACs in matrix using the SCIEX 7500 system
- Sensitive quantification: Achieved a low-pg/mL level LLOQ for the quantification of ARV-110 in rat plasma with minimal sample preparation
- Excellent quantitative performance: Achieve outstanding accuracy, precision and linearity for the analysis of PROTACs using the SCIEX 7500 system featuring improved front-end technology
- Streamlined data management: Increase productivity with a user-friendly interface and integrated platform for data acquisition, processing and management for routine bioanalysis using SCIEX OS software
Methods
Sample preparation: ARV-110 was spiked into 100 μL rat plasma at concentrations ranging from 5 pg/mL to 5000 pg/mL. Protein precipitation was performed using a 600 μL aliquot of 1:1 (v/v) methanol:acetonitrile. Samples were vortexed for 1 min followed by centrifugation at 13,000 rpm for 10 min at 4°C. The supernatant was aliquoted and dried under nitrogen gas. Samples were reconstituted using 200 μL of 1:1 (v/v) methanol:water prior to analysis.
Chromatography: Samples were analyzed using an ExionLC AC system at a flow rate of 0.3 mL/min with a Phenomenex Kinetex C18 (2.1 x 100 mm, 2.6 µm, 100 Å) column and a 10 minute gradient (Table 1). The operating column temperature was 40°C. Mobile phase A was 0.05% formic acid in water and mobile phase B was 0.05% formic acid in acetonitrile. A 10 µL injection volume was used for analysis.
Method development and quantitative performance
Several PROTACs have been validated at the preclinical stage including ARV-110, which reached clinical trials in 2019.3 With several promising PROTAC candidates in the horizon, thorough preclinical and clinical analyses must be performed. To support proper drug product evaluation for safety and efficacy, rapid and sensitive bioanalytical methods are needed for an accurate quantitative determination in matrix.
In this study, the SCIEX 7500 system was employed for the quantification of ARV-110 in rat plasma. The SCIEX 7500 system is equipped with key hardware features that provide significant gains in the generation, capture and transmission of ions. This offers high levels of sensitivity and quantification power for analytes in complex matrices.
A [M+H]+ at m/z 812.4 was selected as the precursor ion of ARV110. The most dominant fragment ion of ARV-110 was at m/z 452.2 and was chosen for quantification. For structural confirmation, a secondary fragment ion at m/z 562.2 was monitored.
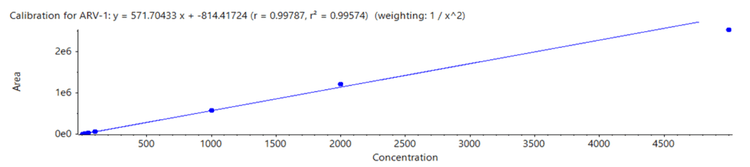
PROTAC concentrations ranging from 5 pg/mL to 5000 pg/mL were analyzed and a LDR spanning 3 orders of magnitude was observed. Strong linearity was reached with a correlation coefficient (r2 ) of 0.996 (Figure 2).
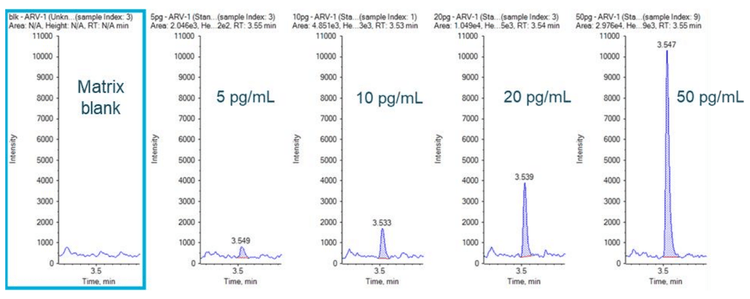
An LLOQ of 5 pg/mL was achieved for ARV-110 in rat plasma with a total run time of 7 mins (Figure 3). No interferences were observed in the matrix blank at the retention time of the compounds, as shown in Figure 3.
The quantification results are summarized in Table 4. Overall, the accuracy was within ±11.2% of the nominal concentration for ARV-110.
Conclusion
- An LLOQ of 5 pg/mL was reached for quantification of ARV110 in rat plasma with minimal sample preparation
- A highly sensitive assay for quantification of PROTACs was demonstrated on the SCIEX 7500 system with an improved front-end technology for better ion generation, capture and transmission
- The method demonstrated excellent accuracy, precision and linearity at all concentration levels
- A single platform for streamlined data acquisition, processing and management with SCIEX OS software is presented
References
- Kathleen M. Sakamoto, Kyung B. Kim, Akiko Kumagai, Frank Mercurio, Craig M. Crews and Raymond J. Deshaies (2001). Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. PNAS 98(15): 8554-8559.
- Mariell Pettersson and Craig M Crews (2009). PROteolysis TArgeting Chimeras (PROTACs) - Past, present and future. Drug Discov Today Technol. 31: 15-27.
- Thi-Thao-Linh Nguyen, Jin Woo Kim, Hae-In Choi, Han-Joo Maeng and Tae-Sung Koo (2022). Development of an LCMS/MS Method for ARV-110, a PROTAC Molecule, and Applications to Pharmacokinetic Studies. Molecules 27(6): 1977



