Abstract
This technical note presents the characterization of mRNA-LNP samples using an optimized sample preparation method developed in a previous study.1 The method eliminates the heat incubation during sample preparation of both mRNA and mRNA-LNP formulations to prevent heat-induced degradation, thereby significantly enhancing the accuracy of mRNA integrity analysis. Following sample preparation, the samples are analyzed using capillary gel electrophoresis with laser-induced fluorescence detection (CGE-LIF), a robust and sensitive technique well-suited for evaluating mRNA integrity, purity, stability, and encapsulation efficiency (EE%) in LNPs.
In this study, the optimized method was further validated and applied to characterize several critical quality attributes (CQAs) of mRNA-LNP samples during their production. First, the repeatability of mRNA integrity analysis was assessed for both mRNA drug substance (DS) and mRNA-LNP drug product (DP), demonstrating excellent consistency and reliability. Second, the method’s quantitation capability was confirmed by establishing linearity across a range of mRNA concentrations, with R² values ≥ 0.994 across six independent runs. Third, a forced degradation study under elevated temperatures was performed to assess mRNA stability, revealing clear temperature-dependent degradation patterns. Finally, the method was applied to determine EE%, achieving an average of 93.8% with high reproducibility.
Overall, this optimized sample preparation and CGE-LIF analysis method provides a reliable, accurate, and comprehensive approach for characterizing mRNA-LNP products, supporting the development, quality control, and regulatory evaluation of high-quality mRNA-based therapeutics and vaccines.
Key features
- Heat-free, optimized sample preparation: Eliminates heat-induced degradation during processing, ensuring accurate and reliable mRNA integrity analysis.
- High precision and reproducibility: Delivers excellent repeatability (RSD < 0.5% for migration time; RSD < 2.5% for purity) and robust quantification (R² ≥ 0.994) across multiple runs.
- Comprehensive characterization: Enables effective assessment of mRNA stability, EE% (average 93.8%), and integrity using a sensitive CGE-LIF platform—supporting quality control of mRNA-LNP therapeutics.
Introduction
mRNA-based therapeutics and vaccines have emerged as a transformative class of biopharmaceuticals, offering rapid development timelines, scalable manufacturing, and flexible design capabilities.2 LNPs are widely used to encapsulate and deliver mRNA in vivo, protecting it from enzymatic degradation and enabling efficient cellular uptake.3 As mRNA-LNP formulations progress through development into clinical and commercial manufacturing, there is a growing need for robust, high-resolution analytical methods to characterize CQAs such as mRNA integrity, purity, stability, and EE%.4, 5
However, the inherently fragile nature of mRNA poses significant analytical challenges, particularly during sample preparation, where conventional heat-based methods can lead to degradation and inaccurate assessments.6 To address this issue, we previously developed an optimized sample preparation protocol that avoids heat incubation and instead utilizes Triton X-100 and formamide with shaking at room temperature. This gentle approach preserves RNA integrity while enabling efficient release of mRNA from LNPs.
In this technical note, we apply and further validate the optimized method for the comprehensive characterization of mRNA-LNP samples using CGE-LIF. CGE-LIF is a highly sensitive and reproducible platform ideally suited for analyzing mRNA size, purity, and degradation profiles. The method is evaluated across key analytical dimensions, including repeatability, quantitation capability, stability under thermal stress, and EE%. Together, these assessments support the robustness, reliability, and applicability of the optimized workflow for the development and quality control of mRNA-based therapeutics and vaccines.
Materials
The RNA 9000 Purity & Integrity kit (P/N: C48231) was from SCIEX (Framingham, MA) and contained Nucleic Acid Extended Range Gel, SYBRTM Green II RNA Gel Stain*, Acid Wash/Regenerating Solution, CE Grade Water and ssRNA Ladder. The BioPhase BFS capillary cartridge - 8 x 30 cm (P/N: 5080121), BioPhase sample and reagent plates (4,4,8) (P/N: 5080311) and sample loading solution (SLS, or formamide, P/N: 608082) were from SCIEX. The 0.2 µm syringe filter (P/N: 4612) was from PALL (Port Washington, NY). Rainin LTS filter tips were from Mettler Toledo (Oakland, CA). Nuclease-free water (NFW, P/N: AM9932) and Surfact-Amps X-100 (10% (v/v) (Triton X-100, P/N: 28314) were obtained from Thermo Fisher Scientific (Waltham, MA). The firefly luciferase (FLuc) mRNA (P/N: L-7602, 1929 nucleotides) was from TriLink BioTechnologies (San Diego, CA). In this technical note, it was used as a model to represent the DS of an mRNA-based vaccine. Moderna-like lipid nanoparticles (LNPs) encapsulating CleanCap® FLuc mRNA, also supplied by TriLink, were formulated by SINTEF (Trondheim, Norway) and used as the model DP for mRNA-LNP vaccine analysis in this technical note.
Sample preparation
Sample preparation for calibration curve: FLuc mRNA was diluted with CE-grade water, and formamide (50% final concentration) was added to each sample to achieve an 8-point calibration curve of mRNA at a concentration from 0.5 to 13.5 µg/mL.
Optimized sample preparation for mRNA DS sample: 10 µL of the mRNA was mixed with 40 µL of CE-grade water. Finally, 50 µL of formamide (50% final concentration) was added, and the sample was incubated at room temperature for 10 min.
Optimized sample preparation for the mRNA-LNP sample: 10 µL of the mRNA-LNP sample was mixed with 15 µL of Triton X-100 to achieve a final concentration of 1.2%. The mixture was shaken at 800 rpm at room temperature (RT) for 10 min. Subsequently, 25 µL of CE-grade water was added and the sample mixed thoroughly. Finally, 50 µL of formamide (50% final concentration) was added, and the sample was incubated at room temperature for 10 min.
Instrument methods
A BioPhase 8800 system (P/N 5083590) equipped with LIF detection was from SCIEX. The excitation and emission wavelengths used were 488 nm and 520 nm, respectively. Data acquisition and analysis were performed using BioPhase software, version 1.2.20.
The conditioning method, separation methods (pressure injection mode or electrokinetic injection mode), and the shutdown method used in this technical note were described in the application guide of the RNA 9000 Purity & Integrity kit.10
Mass spectrometry: The ZenoTOF 7600 system was used with an OptiFlow Turbo V source in either nanoflow or analytical flow mode. SCIEX OS 3.3.1 software was used for instrument control and data acquisition. Source parameters are listed in Table 2. Zeno SWATH DIA data, unless otherwise specified, was acquired using the parameters listed in Table 3. Scan time was 1.6 s for nanoflow-LC-MS and 1.3 s for analytical flow LC-MS, ensuring a minimum of 6 points across the chromatographic peaks.
Data processing: Data was processed using DIA-NN 1.9 software. 3,4 Library-free searches were performed on 5 replicates using a UniProt-reviewed mouse protein database to which the UPS2 human spike-in proteins were added. Search parameters used, unless otherwise specified, are listed in Table 4. Proteins were filtered at a Global Protein Group Q value of 0.01. The Analytics module of SCIEX OS software version 3.3.1 was used for the quantitation of HCPs as well.
Results and discussion
Optimized mRNA and mRNA-LNP sample preparation method: As discussed in the previous technical note,1 the fragile nature of mRNA presents significant challenges during the preparation of samples for mRNA-based therapeutics and vaccines. Heat incubation, commonly used during the preparation of mRNA drug substances and mRNA-LNP drug products, can lead to mRNA degradation, compromising the accuracy of analytical results. To address this, an optimized sample preparation protocol was developed that avoids heat incubation to preserve mRNA integrity.
For mRNA sample analysis, 50% formamide was added to denature the mRNA prior to loading onto the BioPhase 8800 system. For mRNA-LNP samples, the optimized protocol involved treatment with 1.2% Triton X-100 to disrupt the lipid nanoparticles, followed by the addition of formamide and shaking at 800 rpm at room temperature for 10 minutes. This approach maximized mRNA release while minimizing degradation. In addition, pressure injection was identified as the preferred method for sample introduction, particularly for samples with low concentration or high salt content.
The performance of this optimized sample preparation method was further evaluated for the characterization of both mRNA and mRNA-LNP samples. The refined protocol significantly improved the reliability and reproducibility of mRNA integrity analysis, thereby supporting the development of high-quality mRNA-LNP drug products essential for advancing mRNA-based therapeutics and vaccines.
In this technical note, Fluc mRNA was used as a model to demonstrate the optimized method for assessing mRNA integrity, stability (of both mRNA and mRNA-LNPs), and LNP EE% during mRNA-LNP manufacturing.
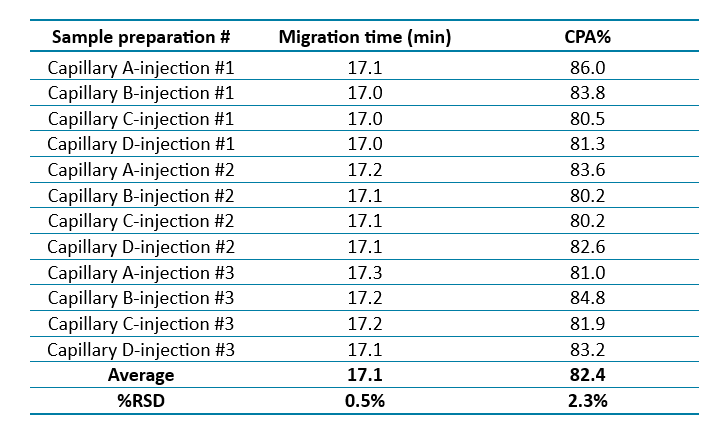
Excellent repeatability of mRNA integrity analysis: To ensure consistent and reliable mRNA integrity analysis, the method's repeatability was evaluated for both mRNA DS and mRNA-LNP DP samples. The results demonstrated excellent repeatability, as indicated by the low %RSD values for both the migration time (MT) of the mRNA main peak and sample purity.
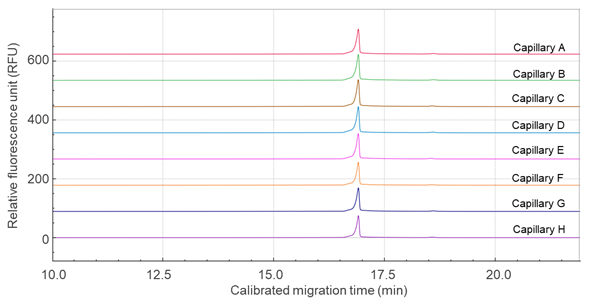
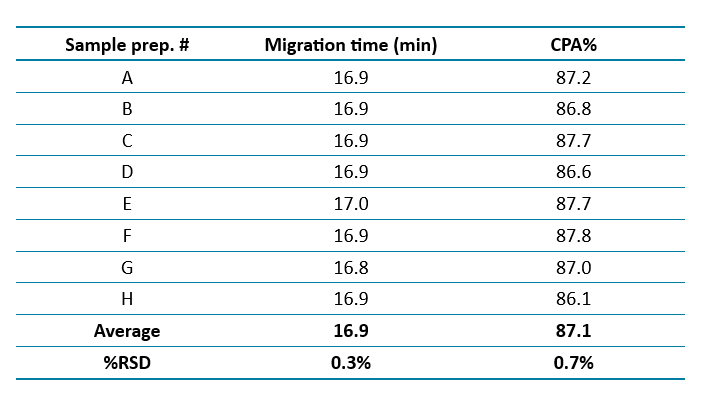
Figure 2 presents the overlaid electropherograms of mRNA DS analyzed across eight capillaries using the multi-capillary BioPhase 8800 system. The calibrated migration time was applied to align the main peak across all eight capillaries. Table 1 summarizes the repeatability data, showing %RSD values of 0.3% for migration time and 0.7% for purity of the mRNA main peak, both indicating high analytical precision.
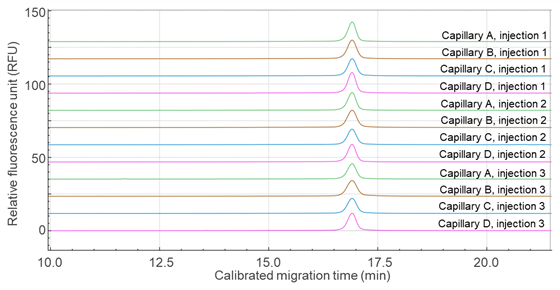
Similarly, Figure 3 shows overlaid electropherograms of the mRNA-LNP drug product analyzed on four different capillaries, each with three replicates. Again, the calibrated migration time was used to align the main peaks across the traces. Table 2 presents the repeatability data for the mRNA-LNP samples, with %RSD values of 0.5% for migration time and 2.3% for purity, confirming the method's robustness and consistency.
Overall, the optimized sample preparation and analysis protocol demonstrates high repeatability and reproducibility for mRNA integrity assessment. This robust method provides a reliable analytical tool for the quality control of both mRNA drug substances and mRNA-LNP drug products.
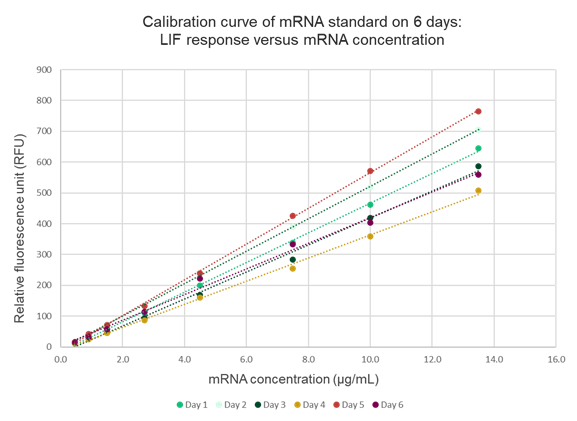
Some variation in calibration curve intensities was observed across the six runs, with approximately 15% variability in signal response between days. This variation can be attributed to several factors. First, the mRNA standards were freshly prepared for each run, potentially introducing slight differences in concentration or handling. Second, minor inconsistencies in the amount of dye added to the separation gel during each preparation may have affected signal intensity. Additionally, detector variability between runs could also contribute to the observed differences. Given these factors, a calibration curve should be freshly prepared alongside the samples for each analytical run to ensure accurate and consistent quantitation.
Overall, this optimized method provides a reliable and robust analytical tool for the quantitation of mRNA in mRNA-LNP samples, supporting the development and quality control of high-quality mRNA-based therapeutics and vaccines.
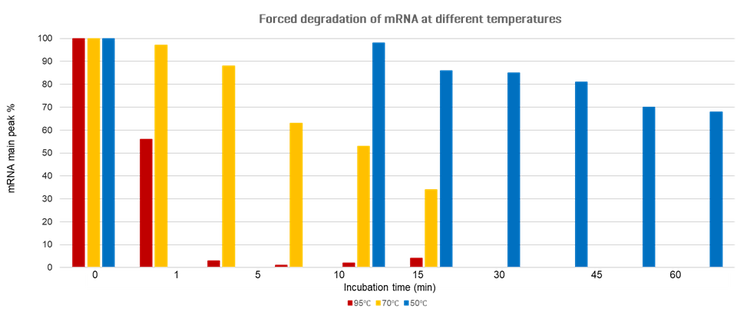
Forced degradation study: To investigate the stability of mRNA-LNP samples under various stress conditions, a forced degradation study was conducted using the optimized method. The samples were subjected to different temperatures (95°C, 70°C, and 50°C) for varying incubation times.
Figure 1 shows the percentage of intact mRNA detected at different incubation times and temperatures, normalized to the 0-minute time point. The plots illustrate the degradation kinetics of mRNA in mRNA-LNP samples under each temperature condition. As expected, higher temperatures accelerated mRNA degradation. At 95°C, degradation was rapid, with a substantial reduction in intact mRNA (red bars in Figure 1). At 70°C, degradation occurred more gradually, showing a progressive decline in the intact mRNA signal (yellow bars). At 50°C, degradation proceeded at the slowest rate, with only a modest decrease in intact mRNA over time (blue bars).
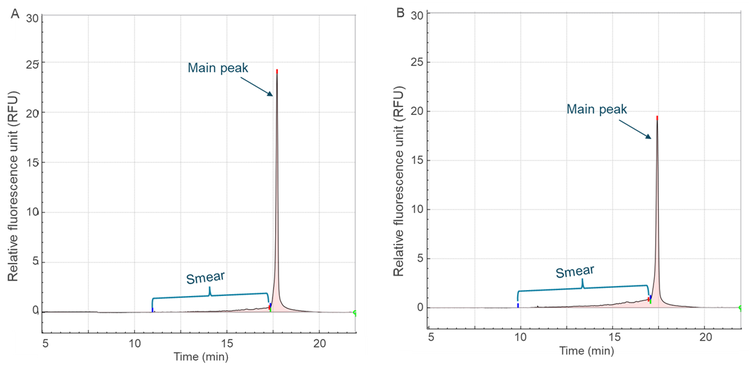
In addition to quantifying the loss of intact mRNA, the extent of degradation was evaluated by measuring the smear signal, representing fragmented RNA. As an example, Figure 5 shows the electropherograms of mRNA-LNP samples incubated at 70°C for 1 minute (Figure 5A) and 30 minutes (Figure 5B). Over time, the intensity of the main peak decreased while the smear signal increased, indicating progressive degradation.
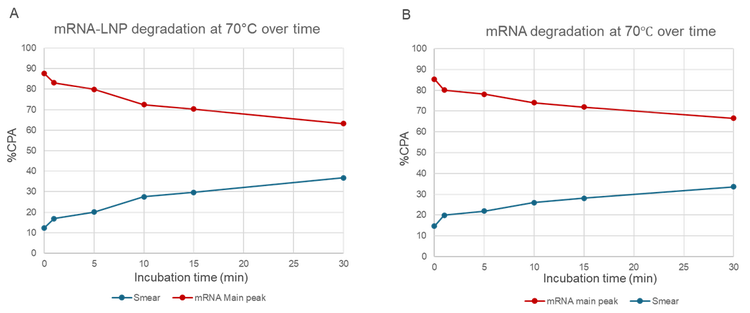
Figure 6 presents a time-course analysis of mRNA degradation at 70°C for both mRNA drug substance (DS) and mRNA drug product (DP) samples. The data demonstrate a consistent decrease in intact mRNA and a corresponding increase in smear signal with longer incubation times, further demonstrating the method's ability to detect and quantify both intact and degraded species.
These results highlight the critical impact of temperature on mRNA stability and underscore the importance of stringent temperature control during the manufacturing, storage, and handling of mRNA and mRNA-LNP products. The optimized method proved effective in monitoring both intact and degraded mRNA, confirming its robustness and reliability for stability studies.
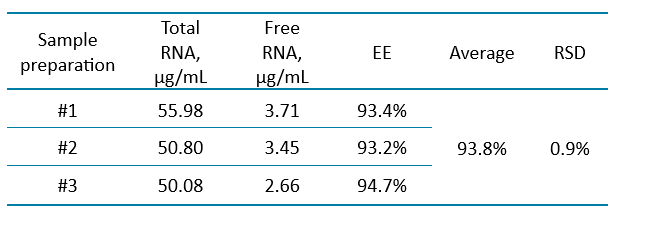
EE% determination: To determine the EE% of the mRNA-LNP drug product, samples and calibration curves were prepared in a 96-well plate format. Two calibration curves were generated: one for the measurement of total mRNA (using Triton X-100 to disrupt the LNPs during sample preparation) and the other for the quantification of free, unencapsulated mRNA. All samples were prepared in triplicate for both free RNA and total RNA analyses.
EE% was calculated based on the difference between the measured total RNA and free RNA concentrations. The results, summarized in Table 3, show an average EE% of 93.8%, with a relative standard deviation (RSD) of 0.9% across three replicates. This high EE% indicates that the vast majority of mRNA was successfully encapsulated within the lipid nanoparticles, minimizing the amount of unencapsulated (free) RNA. The inclusion of Triton X-100 in the sample preparation step effectively disrupted the lipid nanoparticles, enabling accurate quantitation of total mRNA.
These results confirm that the optimized method is reliable and precise for determining EE%, a critical parameter, for evaluating the quality and performance of mRNA-LNP formulations. High EE% ensures effective protection and delivery of mRNA, supporting the development of stable and efficacious mRNA-based therapeutics and vaccines.
Conclusion
In summary, this technical note demonstrates the effectiveness of the optimized release method for characterizing mRNA-LNP samples using capillary gel electrophoresis (CGE). The key findings are:
- Optimized, heat-free sample preparation: Eliminates heat incubation during sample prep to prevent mRNA degradation, significantly improving the accuracy and integrity of mRNA analysis.
- Excellent repeatability and quantitation capability: Ensures consistent and reliable mRNA integrity results for both mRNA drug substances (DS) and mRNA-LNP drug products (DP), with low %RSD in peak migration time and purity across multiple capillaries.
- Outstanding quantitation capability: Demonstrates excellent linearity (R² ≥ 0.994) across a wide range of mRNA concentrations over six independent runs, enabling accurate and reproducible quantitation.
- Robust forced degradation assessment: Evaluates mRNA and mRNA-LNP stability under thermal stress (95°C, 70°C, 50°C), highlighting temperature-dependent degradation patterns and the method’s sensitivity in detecting both intact and degraded species.
- Accurate EE% determination: Measures the total and free RNA to determine EE%, achieving an average EE of 93.8% with high precision and reproducibility.
- Comprehensive characterization: The optimized method supports the development of high-quality mRNA-LNP products, essential for advancing mRNA-based therapeutics and vaccines.
References
- mRNA release improvement for mRNA integrity analysis in mRNA -LNP drug product by Capillary Electrophoresis (CE). SCIEX technical note, MKT-34984-A.
- Pardi, N. et al. mRNA vaccines for infectious diseases - advances, challenges and opportunities. Nat Rev Drug Discov. 2024 Nov;23(11):838-86.
- Hou, X. et al. Lipid nanoparticles for mRNA delivery. Nat Rev Mater 2021 6, 1078–1094.
- Huang, X. et al. Unlocking the Therapeutic Applicability of LNP-mRNA: Chemistry, Formulation, and Clinical Strategies. Research (Wash D C). 2024 Jun 18;7:0370.
- Leighton, L.J. et al. The design, manufacture and LNP formulation of mRNA for research use. Nat Protoc 2025.
- Kornienko IV et al. RNA Stability: A Review of the Role of Structural Features and Environmental Conditions. Molecules. 2024 Dec 18;29(24):5978.

