Abstract
This technical note details the optimization of the messenger RNA (mRNA) release method for mRNA-lipid nanoparticle (mRNA-LNP) samples to enhance mRNA integrity analysis using CGE. CGE is a robust and suitable technique for assessing mRNA integrity, purity, stability, and encapsulation efficiency in LNPs.1, 2 Ensuring mRNA integrity is crucial to produce high-quality mRNA-LNP products. 3 However, the fragile nature of mRNA poses challenges during sample preparation, often resulting in degradation and inaccurate analysis.4 This study aims to optimize the release of mRNA from mRNA-LNP drug products while preserving its integrity. The optimization process evaluated three key factors: the use of Triton X-100 to facilitate the release of encapsulated mRNA, the application of formamide to denature mRNA, and the incubation temperature during formamide treatment.5, 6, 7 Additionally, the injection mode for sample analysis using the CGE technique was assessed. The optimized protocol, involving treatment with 1.2% Triton X-100, followed by the addition of formamide and shaking at room temperature for 10 minutes, ensures maximal mRNA release while minimizing its degradation during the sample preparation procedure. The preferred pressure injection method is effective for low-concentration or high-salt samples.8
This refined sample preparation method enhances the reliability and robustness of mRNA integrity analysis in mRNA-LNP drug products. It supports the development of high-quality mRNA-LNP products, which are essential for advancing mRNA-based therapeutics and vaccines.
Key features
- Optimized mRNA release method:Ensures accurate mRNA integrity analysis of mRNA-LNP samples, minimizing sample preparation-related impurities and artifacts
- Refined injection conditions:Pressure injection is suitable for low-concentration or high-salt samples, providing accurate and reliable results
- Consistent and repeatable results:Demonstrates <1% RSD in both migration time and sample purity
- High linearity for quantification: Offers excellent quantification capabilities with an R² value greater than 0.99, ensuring precise and reliable measurements with superior accuracy and recovery.
Introduction
The integrity of mRNA is a critical factor in the production of high-quality mRNA-LNP products, which are increasingly important in therapeutic applications.3 However, assessing the integrity, purity, stability, and encapsulation efficiency of mRNA within LNPs presents significant challenges due to the inherent heterogeneity and length of the RNA molecules. CE has emerged as a powerful tool for the quality control of therapeutic RNA-based molecules, offering highly efficient separation across a wide size range of RNA.1, 2, 3
Currently, a commonly used approach for RNA extraction from LNPs for characterization involves treatment with Triton X-100, followed by the addition of formamide and the application of heat (70 °C for 3-10 minutes).5, 6, 7, 9 This method leverages formamide to disrupt higher-order structures, thereby enhancing the signal of the main species of nucleic acid in CE analysis. Despite its efficacy, the use of heat during this process raises concerns due to the fragile nature of RNA, which is prone to temperature-dependent degradation.
Given these challenges, there is a pressing need to explore alternative, efficient protocols to extract mRNA from mRNA-LNP complexes without compromising RNA quality. This technical note presents an optimized method for mRNA release from mRNA-LNP samples, aimed at improving the integrity analysis of mRNA using CE. Our study evaluates the use of Triton X-100 to facilitate the release of encapsulated mRNA, the application of formamide to denature RNA, and the incubation temperature during formamide treatment. The optimized protocol includes treatment with 1.2% Triton X-100, followed by the addition of formamide and a 10-minute shaking at room temperature. This approach maximizes mRNA release while minimizing degradation during sample preparation. This refined sample preparation method significantly enhances the reliability and reproducibility of mRNA integrity analysis, addressing the critical need for efficient protocols that do not compromise RNA quality. Additionally, the preferred pressure injection method proves effective for samples with low concentration or high salt content.
Methods
Materials: The RNA 9000 Purity & Integrity kit (P/N: C48231) was from SCIEX (Framingham, MA) and contained Nucleic Acid Extended Range Gel, SYBRTM Green II RNA Gel Stain*, Acid Wash/Regenerating Solution, CE Grade Water and ssRNA Ladder. The BioPhase BFS capillary cartridge - 8 x 30 cm (P/N: 5080121), BioPhase sample and reagent plates (4,4,8) (P/N: 5080311) and sample loading solution (SLS, or formamide, P/N: 608082) were from SCIEX. The 0.2 µm syringe filter (P/N: 4612) was from PALL (Port Washington, NY). Rainin LTS filter tips were from Mettler Toledo (Oakland, CA). Nuclease-free water (NFW, P/N: AM9932) and Surfact-Amps X-100 (10% (v/v) (Triton X-100, P/N: 28314) were obtained from Thermo Fisher Scientific (Waltham, MA). The firefly luciferase (FLuc) mRNA (P/N: L-7602, 1929 nucleotides) was from TriLink BioTechnologies (San Diego, CA). The Moderna-like LNPs loaded with CleanCap® FLuc mRNA from TriLink were formulated by SINTEF (Trondheim, NO).
Instrument methods: A BioPhase 8800 system (P/N 5083590) equipped with LIF detection was from SCIEX. The excitation and emission wavelengths used were 488 nm and 520 nm, respectively. Data acquisition and analysis were performed using BioPhase software, version 1.2.20.
The conditioning method, separation methods (pressure injection mode or electrokinetic injection mode), and the shutdown method used in this technical note were described in the application guide of the RNA 9000 Purity & Integrity kit.10
Sample preparation:
Sample preparation for the temperature influence experiment for the mRNA sample: 5 µL of FLuc mRNA was mixed with 15 µL of CE-grade water and 50 µL of formamide and incubated at 70 °C using a thermal cycler for 0, 1, 5, 10, 15, and 30 minutes (min). Then, samples were chilled on ice for 5 min, and 30 µL of CE-grade water was added before loading onto the BioPhase 8800 system for analysis. The final concentration of formamide was 50%.
Sample preparation for the temperature influence experiment for the mRNA-LNP sample: 10 µL of mRNA-LNP sample was mixed with 15 µL of Triton X-100, incubated at room temperature for 15 minutes, and 50 µL of formamide and incubated at 70 °C using a thermal cycler for 0, 1, 5, 10, 15, and 30 min. Then, samples were chilled on ice for 5 minutes, and 25 µL of CE-grade water was added before loading onto the BioPhase 8800 system for analysis. The final concentration of formamide was 50%.
Sample preparation for calibration curve: FLuc mRNA was diluted with CE-grade water, and formamide (50% final concentration) was added to each sample to achieve an 8-point calibration curve of mRNA at a concentration from 0 to 15 µg/mL.
Optimized sample preparation for the mRNA-LNP sample: 10 µL of the mRNA-LNP sample was mixed with 15 µL of Triton X-100 to achieve a final concentration of 1.2%. The mixture was shaken at 800 rpm at room temperature (RT) for 10 min. Subsequently, 25 µL of CE-grade water was added and mixed thoroughly. Finally, 50 µL of formamide (50% final concentration) was added and incubated at room temperature for 10 min. Instrument
Results and discussion
Influence of heat incubation during sample preparation on mRNA degradation: In this study, we investigated the impact of 70°C incubation, a commonly used temperature during sample preparation for mRNA integrity analysis, on the degradation of mRNA, both in its free form and encapsulated within lipid nanoparticles (LNPs), as shown in Figure 2. The analysis was conducted using the SCIEX multi-capillary system, with samples incubated at 70°C for varying durations (0, 1, 5, 10, 15, and 30 min).
For mRNA drug substances, Figure 2A illustrates the percentage of mRNA degradation over time when incubated at 70°C. The y-axis represents the percentage of intact mRNA remaining after the heat incubation, while the x-axis indicates the incubation time. The graph demonstrates a rapid increase in mRNA degradation, starting from 4% of mRNA degraded after 1 minute of heating, 16% after 10 minutes, and reaching 34% after 30 minutes of incubation. These results underscore the sensitivity of mRNA to thermal stress and the significant impact of high-temperature incubation on mRNA integrity.
Similarly, mRNA encapsulated within LNPs exhibited a comparable degradation pattern, as shown in Figure 2B. The mRNA extracted from these LNP samples showed noticeable degradation after only 5 minutes of incubation at 70°C. After 30 minutes, nearly 50% of the initial mRNA was degraded. This substantial degradation underscores the vulnerability of mRNA to high temperatures when releasing the mRNA from the LNPs.
These observations suggest that sample preparation involving mRNA should avoid heat incubation due to its detrimental effects on mRNA integrity. The study emphasizes the necessity for further exploration of alternative, efficient protocols that can prepare mRNA for integrity analysis without compromising its quality. Future research should focus on identifying and optimizing conditions that minimize mRNA degradation during sample preparation, thereby preserving the integrity and providing accurate results of the integrity analysis.
In conclusion, the study highlights the need for further exploration of alternative, efficient protocols capable of extracting fragile mRNA from mRNA-LNPs without inducing sample preparation-related degradation, which would result in inaccurate integrity analysis. To minimize or avoid sample preparation-related RNA degradation, incubation with formamide at RT with shaking at 800 rpm, instead of any heat incubation, will be used moving forward in the study for further optimization. This approach is expected to better preserve mRNA integrity and enhance the reliability of mRNA analysis.
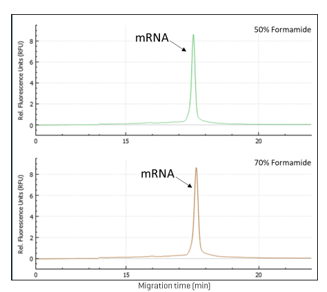
Optimization of formamide used for the sample preparation: This study optimized the use of formamide in the sample preparation process for mRNA analysis. Two concentrations of formamide, 50% and 70%, were evaluated using the SCIEX multi-capillary system and the RNA 9000 Purity & Integrity kit. The results are presented in Figure 4 (50% formamide vs 70% formamide).
Our analysis revealed no significant differences in peak shape and peak intensity between the two formamide concentrations. This indicates that both 50% and 70% formamide are equally effective for mRNA sample preparation under the conditions tested. The consistency in peak characteristics suggests that either concentration can be used without compromising the quality of the RNA analysis.
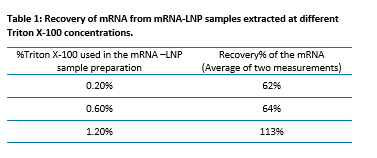
Optimization of Triton X-100 used for the sample preparation of the mRNA-LNP drug product: In this study, we optimized the use of Triton X-100 detergent for the sample preparation of mRNA-LNP drug products. The same mRNA-LNP sample was treated with three different concentrations of Triton X-100 (final concentrations of 0.2%, 0.6%, and 1.2%) for 15 min at RT. Each sample preparation was duplicated, and the samples prepared were analyzed using the RNA 9000 Purity & Integrity kit and the BioPhase 8800 system. The concentration and recovery of the mRNA extracted from the mRNA-LNP samples were calculated based on the calibration curve of mRNA of known concentration.
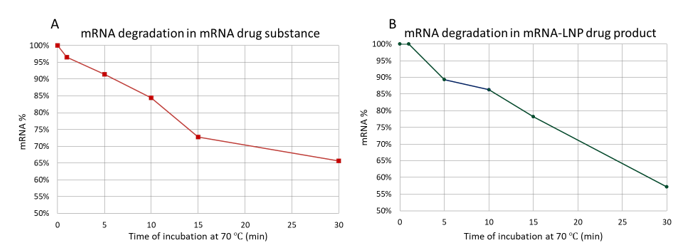
Table 1 shows the recovery of mRNA extracted from the mRNA-LNP samples at different Triton X-100 concentrations, shaken at 800 rpm for 15 minutes, followed by the addition of formamide to a final concentration of 50%. The recovery is the ratio of the total mRNA in the sample and the mRNA loaded when the LNPs are prepared. Recovery over 100% can be obtained due to the matrix effects generated by lipids present in the sample but not in the calibration curve.
These findings suggest that using a concentration of Triton X-100 of 1.2% is effective for extracting mRNA from LNPs while preserving the integrity of the mRNA. This optimized protocol provides a reliable approach for preparing mRNA-LNP samples for analysis, ensuring high recovery and minimal degradation.
Future research should explore the impact of alternative detergents and shaking times on mRNA extraction efficiency and stability. Additionally, further studies could investigate the compatibility of this optimized protocol with various mRNA-LNP formulations to ensure its broad applicability across different lipid chemistries.
Overall, optimizing Triton X-100 concentration is a critical step in refining sample preparation methods for mRNA-LNP drug products, contributing to more accurate and reproducible results in mRNA analysis.
Injection Modes for mRNA Integrity Analysis: In this study, we evaluated the effectiveness of two different injection modes for mRNA integrity analysis using the RNA 9000 Purity & Integrity kit on the BioPhase 8800 system. An 8-point calibration curve was prepared with mRNA concentrations ranging from 0.5 µg/mL to 15 µg/mL. The same set of calibration curves was analyzed using both electrokinetic and pressure injection modes.
The results, presented in Figure 5, indicate that the electrokinetic injection mode produced lower signal intensities for mRNA compared to the pressure injection mode. This discrepancy is attributed to the competition of matrix components in the sample solution during electrokinetic injection, which can affect the mRNA signal.
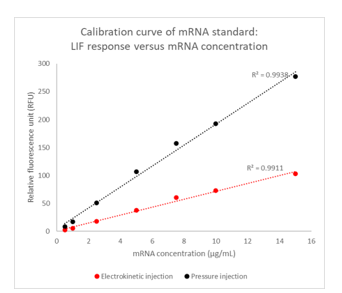
Given these findings, the pressure injection is preferred for analyzing low-concentration samples or in-process/crude samples that contain higher salt concentrations. The pressure injection mode minimizes the impact of matrix competition, resulting in more accurate and reliable mRNA integrity analysis. During electrokinetic injections, samples compete with the salt present in the solution, leading to reduced samples being injected for analysis. Consequently, for samples with higher salt content, pressure injection provides a more robust and consistent analysis, ensuring the integrity and accuracy of the results.
Repeatability of mRNA analysis of mRNA-LNP samples using the optimized sample preparation protocol: In this study, we evaluated the repeatability of mRNA integrity analysis for mRNA-LNP drug products using an optimized sample preparation and analysis protocol. The protocol involved incubating the mRNA-LNPs in Triton X-100 (1.2% v/v) for 15 minutes with shaking at 800 rpm. Subsequently, 50% formamide was added without heating. The samples were then injected using standard pressure injection with the RNA 9000 Purity & Integrity kit and analyzed on the BioPhase 8800 system.
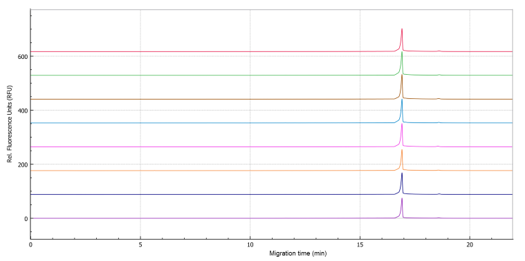
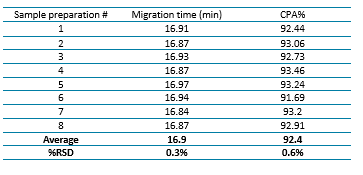
Figure 1 shows the stacked traces of 8 sample preparations analyzed across 8 capillaries on the BioPhase 8800 system. The relative standard deviations (RSDs) for migration time and corrected peak area percentage (CPA%) for these analyses are presented in Table 2. The results demonstrate <1% RSD in both the main peak migration time and mRNA integrity analysis, indicating a high level of consistency and reliability in the mRNA integrity analysis.
These findings demonstrate that the optimized sample preparation and analysis protocol provides a robust and repeatable method for assessing mRNA integrity in mRNA-LNP samples. The protocol involves mixing 10 µL of the mRNA-LNP sample with 15 µL of 2% Triton X-100 (1.2% final concentration), shaking at 800 rpm at room temperature for 10 minutes, adding 25 µL of CE-grade water, and finally adding 50 µL of formamide (50% final concentration) and incubating at room temperature for 10 minutes. The use of Triton X-100 and formamide without high-temperature heat incubation in the preparation process effectively preserves mRNA integrity, while the BioPhase 8800 system ensures accurate and reproducible analysis.
Future studies will be conducted to continue refining and validating this protocol across different mRNA formulations and experimental conditions to further enhance its applicability and reliability in mRNA-based therapeutic development.
Conclusion
This technical note underscores the importance of optimized sample preparation and analysis protocols to ensure the accuracy and reliability of mRNA integrity analysis in mRNA-LNP samples, enhancing the development of mRNA-based therapeutics.
- Optimized Method: This method provides accurate mRNA integrity analysis of mRNA-LNP samples, reducing impurities and artifacts during sample preparation.
- Flexibility in Injection Mode: Pressure injection is preferred for samples with low concentration or high salt content, enhancing the reliability and accuracy of results.
- Consistent and reproducible performance: Both migration time and sample purity exhibit less than 1% RSD, guaranteeing reliable and repeatable outcomes.
- Exceptional quantitation linearity: Demonstrates outstanding precision in quantitation, with an R² greater than 0.99, ensuring accuracy.
These optimized protocols are crucial for advancing mRNA-based therapeutics by ensuring high-quality and reliable mRNA integrity analysis.
References
1. Sara S. Nogueira, et al., (2024) Analytical techniques for the characterization of nanoparticles for mRNA delivery, European Journal of Pharmaceutics and Biopharmaceutics. Volume 198, 114235,
2. Tran, J.P., et al., (2025) A Comprehensive Evaluation of Analytical Method Parameters Critical to the Reliable Assessment of Therapeutic mRNA Integrity by Capillary Gel Electrophoresis. Electrophoresis, 46, 365-375.
3. Linde Schoenmaker, et al., (2021) mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability, International Journal of Pharmaceutics, Volume 601, 120586.
4. Uddin MN, et al., (2021) Challenges of Storage and Stability of mRNA-Based COVID-19 Vaccines, Vaccines (Basel), 9(9):1033.
5. Camille Malburet, et al., (2024) mRNA extraction from lipid nanoparticles, Journal of Chromatography A, Volume 1714, 464545.
6. Tomas Masek, et al., (2005) Denaturing RNA electrophoresis in TAE agarose gels, Analytical Biochemistry, Volume 336, Issue 1, 46-50.
7. Avci-Adali M, et al., (2014) In vitro synthesis of modified mRNA for induction of protein expression in human cells, J Vis Exp., 93, 51943.
8. Ferreira Santos MS, et al., (2023) A hydrodynamic injection approach for capillary electrophoresis using rotor-stator valves, Electrophoresis, 44(9-10):784-792.
9. A new approach to determine encapsulation efficiency of mRNA-lipid nanoparticles (mRNA-LNP) by capillary gel electrophoresis with laser-induced fluorescence detection. SCIEX technical note. MKT-28376-A.
10. RNA 9000 Purity & Integrity kit for the BioPhase 8800 system, application guide. RUO-IDV-05-13438-B.