Abstract
Improved selectivity and greater sensitivity were achieved for peptide quantitfication in a complex matrix using the ZenoTOF 7600 system featuring the Zeno trap. The greater mass resolution provides selectivity between target peptide versus matrix-derived interferences and general high background effects. This enhanced selectivity aids in superior quantitative results in terms of LOQ, precise data integration, improved reproducibility, and linear dynamic range. In addition, enhanced sensitivity for peptide quantification was achievable using the Zeno trap to improve the duty cycle versus traditional time-of-flight systems.
Traditional workflows for quantitative bioanalyses, such as immunological assays, have been displaced by LC-MS/MS analysis on a triple quadrupole mass spectrometer. Immunoassays often lack selectivity, specificity, and have a limited linear dynamic range. While the triple quadrupole platform provides excellent sensitivity and quantitative performance, there can be some limitations with background interference based on the lower resolution considering the type of mass analyzer. Background interference is a common issue for workflows where analytes are present in a highly complex matrix. High-resolution accurate mass spectrometry (HRAMS) has been increasingly adopted for quantitative bioanalysis.1,2 The ZenoTOF 7600 system offers an exceptional combination of mass resolution, sensitivity, and acquisition speed for quantitative analysis. It also aids in more accurate and automated integration, the potential for less ion path tuning, the ability to change measured fragments post-acquisition, and improved reproducibility and LDR when interferences are mitigated. These attributes complement the excellent sensitivity of a nominal mass triple quadrupole system such as the SCIEX Triple Quad 7500 LCMS/MS system − QTRAP Ready or SCIEX Triple Quad 6500+ LC-MS/MS system for a biopharmaceutical lab which requires a full range of capabilities.
The ZenoTOF 7600 system offers a high-resolution MS/MSbased acquisition mode for peptide quantification with Zeno MRMHR, along with improved sensitivity using the Zeno trap. The implementation of the Zeno trap allows for improvement of the duty cycle by ≥90 %, thereby improving overall MS/MS sensitivity.
Key features of the ZenoTOF 7600 system for peptide quantification
- Gain higher sensitivity for peptide quantification using the Zeno trap, by enhancing duty cycle through an accumulation of ions during each TOF pulse
- Achieve greater selectivity between target peptides and matrix-related components with the higher mass resolution offered by the ZenoTOF 7600 system
- Ensure exceptional accuracy and precision for quantitative workflows using the ZenoTOF 7600 system
- Achieve superior sensitivity for peptide quantification using HRAMS MS/MS in comparison with single MS mode
- Perform highly automated and accurate peak integration given the improvements in selectivity with higher resolution and ensure overall data integrity
- Easily acquire, process, and manage data on a single platform using the SCIEX OS software
Methods
Samples and reagents: Universal Proteomics Standard (UPS) was purchased from Sigma-Aldrich. Rat plasma (Sprague Dawley, K2 EDTA) was purchased from BioIVT.
Sample preparation: The calibration curve was prepared by spiking digested UPS into rat plasma digest followed by serial dilution.
Samples were denatured by incubating with N-octyl-glucoside (OGS), followed by reduction with dithiothreitol (DTT) and alkylation with iodoacetamide (IAM). A trypsin/Lys-C digestion was performed at 37 ºC overnight, with an enzyme-protein ratio of 1:25. Formic acid was spiked into the samples to abort digestion. The samples were centrifuged at a speed of 12,000 g and the supernatant was then injected for LC-MS analysis.
Proteins used for this study had limited starting concentrations. Therefore, the final LDRs were narrow for the peptides analyzed.
Chromatography: An ExionLC system was used for analyte separation. A volume of 20 µL was injected for analysis. Mobile phase A consisted of water with 0.1% FA in water, while organic phase B was composed of 0.1% FA in acetonitrile. For analyte separation, the operating flow rate was set to 0.5 mL/min using a Phenomenex Kinetex C18 column (3 x 50 mm, 2.6 µm, 100 Å). The column oven temperature was set to 40 ºC. Chromatographic conditions for analyte separation are shown in Table 1.
Mass spectrometry: Samples were analyzed in triplicate. Method details such as source and gas parameters and MS conditions are summarized in Table 2. Sample analysis was performed using scheduled Zeno MRMHR on the ZenoTOF 7600 system. The ZenoTOF 7600 system provides a scan speed of 133 Hz.
Data processing: MRM data were processed using SCIEX OS 2.0 software. Integration was performed using the MQ4 algorithm. Linear regression with 1/x weighting was used for quantification of all peptides. The XIC peak width was set to 0.05 Da for both MS/MS and MS1 quantification.
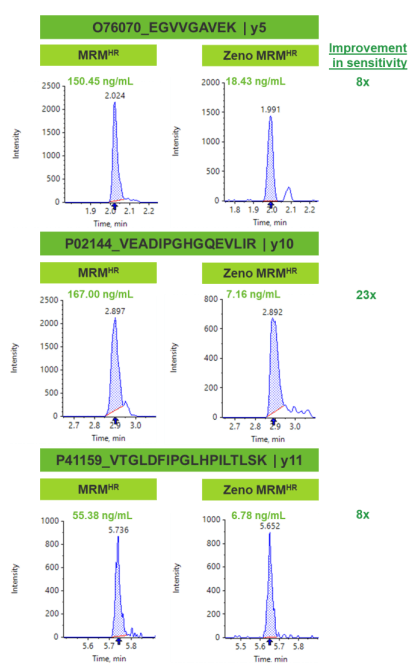
Zeno trap provides greater sensitivity
In traditional time-of-flight MS/MS acquisition, ions are often lost in between TOF pulses due to differences in velocity, with typical duty cycle values range approximately between 5-25%.
Due to loss in ion transmission, there is a subsequent decline in overall sensitivity as fewer ions arrive at the detector. The Zeno trap gains back the ion transmission by providing control of the ion beam from the collision cell into the TOF accelerator (Figure 2). Ions exit the Zeno trap in an ordered release based on potential energy.
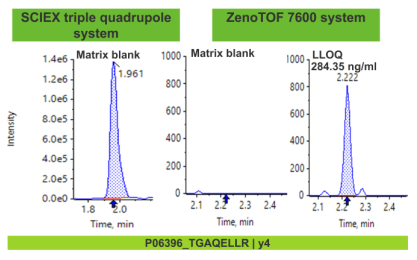
With the design of the Zeno trap, significant enhancements in MS/MS sensitivity were observed. As a result, considerable improvements in LLOQ levels were achieved using Zeno MRMHR in comparison with standard MRMHR (Figure 3). From the three peptide examples shown in Figure 3, greater than 8 times improvement in LLOQ sensitivity was observed.
Selectivity enhancement for peptide quantification
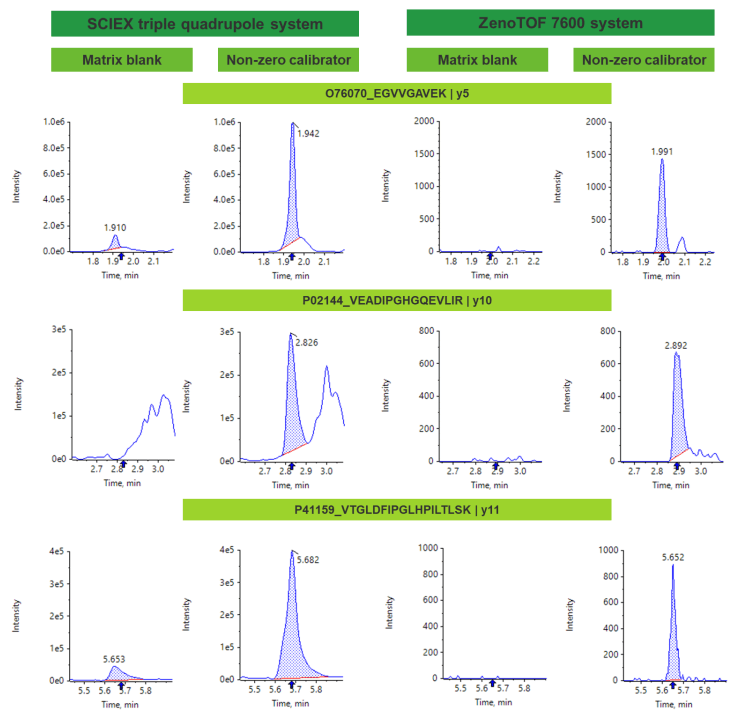
Selectivity issues often arise in quantification workflows in the form of matrix interference and high background. Matrix interference is the presence of a defined peak in the matrix blank at the retention time of the analyte where the precursor and fragment ions are the same as that of the target analyte. High background arises as a result of fragment ions that elute at the retention time of the analyte, which often limits the ability to reach trace levels of quantification.
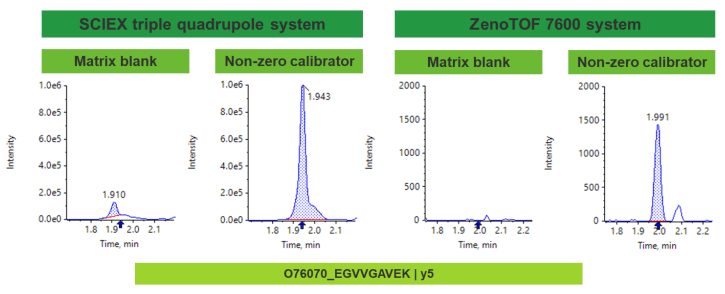
Higher resolution mass spectrometers can maximize selectivity by offering a greater mass resolution between target analyte and any matrix-related components or background ions. With the high resolution capabilities of the ZenoTOF 7600 system, common bioanalysis challenges with matrix interference and high background can be mitigated in comparison with a nominal mass system (Figure 4).
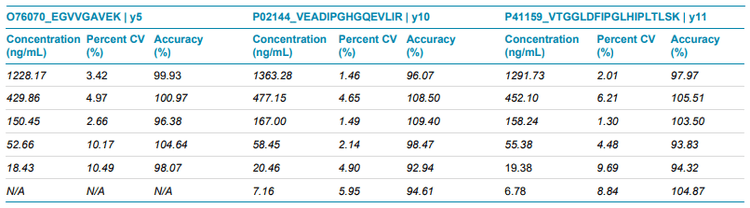
The ZenoTOF 7600 system also achieved strong linearity, accuracy and precision, demonstrating GLP-level quantitative performance (Figure 4, Table 3). Accuracy at the LLOQ was within 80%-120%, while accuracy for all other non-zero calibrators were within 85%-115% of the nominal concentration. Overall, precision was <11%, demonstrating high reproducibility.
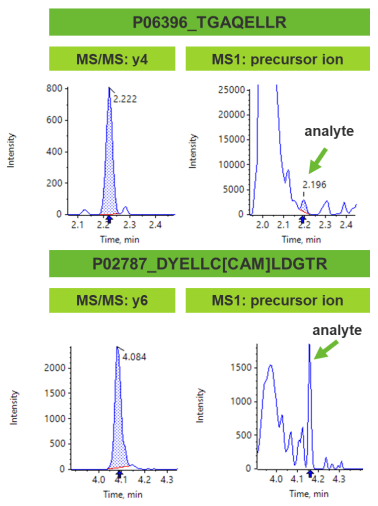
Comparison between MS/MS and MS1 based quantification
In addition to the MS/MS information for MRMHR analysis, quantification can also be performed using the precursor ion associated with the target peptide. Implementation of the precursor ion for quantification does not include additional confirmation from any fragment ions to verify the measured peptide structure. Mainly, when the peptide is present in a challenging matrix, precursors might be present that do not correspond to the target peptide, which may hinder quantification by introducing interferences. As a result, the selectivity of the target peptide can be compromised.
In comparison with MS/MS quantification, MS1 based quantification lacks additional selectivity, causing the S/N to often be lower (Figure 6). When MS/MS data is applied for quantification, S/N is typically considerably enhanced with the fragment ion data providing greater selectivity of the target peptide. Therefore, for peptides present in a complex matrix, MS/MS-based quantification offers a highly selective and specific approach.
Automated and accurate peak integration
Efficiency of integration can often be driven by the presence or absence of matrix interference or high background ions. With a high mass resolution system, target peptide and any matrix interferences and background ions can be easily resolved by mass. With lack of interferences, an XIC with greater selectivity was generated, allowing for automated and accurate peak integration. As shown in Figure 7, co-eluting interferences are separated by greater mass resolution on the ZenoTOF 7600 system when compared to a standard triple quadrupole system.
For bioanalysis workflows, this can be of great advantage, as it allows for more convenient and efficient peak integration. Overall, data integrity can also be enhanced because there is less of a need for manual integration given the greater selectivity provided by high mass resolution.
Conclusions
- Higher sensitivity, based on LLOQ levels, was achieved for peptide quantification using Zeno trap by improving duty cycle through the accumulation of ions during each TOF pulse for enhanced duty cycle
- Greater selectivity was reached between target peptides and matrix-related components with the higher mass resolution offered by the ZenoTOF 7600 system
- GLP compliant accuracy and precision was observed on the ZenoTOF 7600 system, demonstrating GLP-level quantitative performance
- Significantly better S/N was observed for quantification at the MS/MS level in comparison to the MS1 level
- Automated and accurate peak integration was easily attainable on the ZenoTOF 7600 system, with greater mass resolution ensuring overall data integrity
References
- Mike-Qingtao Huang, Zhongping (John) Lin, Naidong Weng (2013). Applications of high-resolution MS in bioanalysis. Bioanalysis 5(10):1269-1276.
- Yuan-Qing Xia, Jim Lau, Timothy Olah, Mohammed Jemal (2011). Targeted quantitative bioanalysis in plasma using liquid chromatography/high‐resolution accurate mass spectrometry: an evaluation of global selectivity as a function of mass resolving power and extraction window, with comparison of centroid and profile modes. Rapid Communications in Mass Spectrometry 25(19):2863-2878.


