Abstract
Data independent acquisition (DIA) has been a central catalyst for shifting mass spectrometry (MS)-based omics from a discovery-focused science to one that more comprehensively facilitates longitudinal studies, as required in clinical research and precision medicine. The challenges associated with improving throughput in high-resolution MS while maximizing depth, reproducibility, and quantitative precision, have given rise to ZT Scan DIA.
ZT Scan DIA comes as SCIEX next iteration of DIA development, following SWATH DIA and most recently, Zeno SWATH DIA. Since SCIEX initial launch of SWATH DIA in 2011, data-independent acquisition increasingly became the method of choice among researchers utilizing liquid chromatography tandem mass spectrometry (LC-MS/MS). SWATH DIA emerged as an LC-MS/MS workflow that capitalizes on the speed and sensitivity gains achievable with modern MS systems. SWATH DIA enables identification of vast numbers of proteins, peptides and metabolites in a more comprehensive manner than conventional data dependent acquisition (DDA) methods. With SWATH DIA, high-quality, high-resolution MS/MS data not only provide identification, but are also the secret behind accurate and precise quantitation.
The development of Zeno trap1,2, a novel accumulation and pulsing device, enabled a new era of sensitivity for accurate mass instruments. In Zeno SWATH DIA, the Zeno trap, when activated, is used to increase the MS/MS sensitivity for each variable window acquired.3 The Zeno trap provides a 4-20x gain in sensitivity for Zeno SWATH DIA, while also maintaining other key performance attributes.4
In this technical overview we introduce a novel DIA approach, ZT Scan DIA, which combines the depth of DIA methods with the precision of targeted approaches. Like Zeno SWATH DIA, ZT Scan DIA enables the identification and quantitation of analytes using MS/MS data. Thus, the rapid acquisition of high-quality MS/MS data across the entire precursor ion space is fundamental to operation. With the added dimension of data from quadrupole scanning, ZT Scan DIA allows researchers to unravel the complexity of biological samples by extending depth and certainty in the quantitative measurement.
Key innovations of ZT Scan DIA
- The powerful combination of DIA, Zeno trap and added specificity of the quadrupole dimension enhances depth and certainty in the quantitative measurements
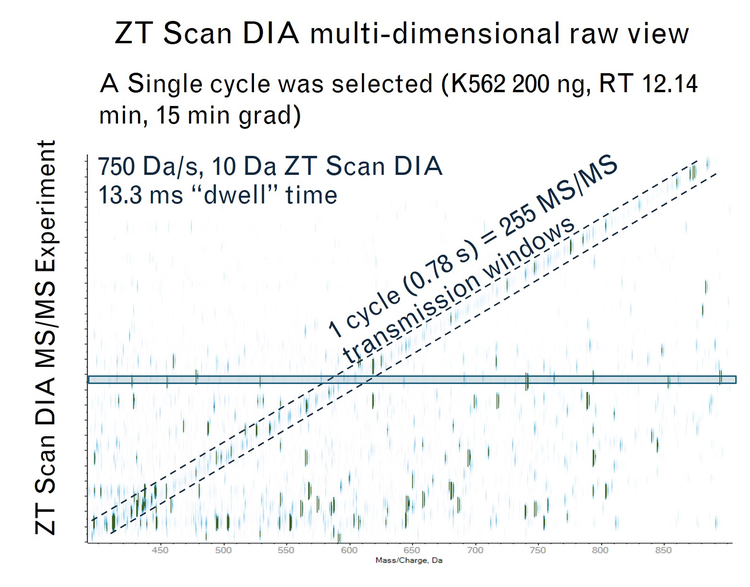
- During each cycle, the isolation window for MS/MS is sliding along the m/z range of interest. Based on the 400-900 m/z range method, scanning at 750 Da/s with a sliding isolation window of 5 Da, this would equate to a scan rate of 640 Hz
- For larger cohorts this can allow up to ten times faster throughput than conventional DIA approaches without compromising data quality
Why do we need DIA?
LC-MS/MS is widely used for characterizing mixtures of unknown compounds. In the conventional approach, DDA is used for acquiring MS/MS data on as many compounds as possible. DDA is well-established and in use for many applications, as it provides an untargeted sample analysis. One major drawback of DDA is that datasets can be incomplete since only precursor ions that match specific criteria are selected for MS/MS analysis. Additionally, detection reproducibility can suffer. Small shifts in retention times from run to run can affect the population of precursor ions entering the instrument during each cycle, and therefore the subset of compounds that are analyzed. Despite recent advances in MS technology performance, limitations remain when attempting to characterize compounds in highly complex matrices. To counter this, a target list of precursors can be used to focus the analysis on desired analytes, however this negates the benefit of a truly unbiased and global approach. Therefore, when comparing multiple DDA data sets, missing peaks and gaps are often observed, which can negatively impact the detection of lower-level analytes.
With DDA, quantification can be performed at the MS1 level using label-free approaches or at the MS2 level using labelling techniques, with the latter requiring additional sample preparation protocols and cost. For this reason, DDA analyses typically use the intact precursor masses found in MS full scan data for quantification, and often identification. Co-eluting isobaric analytes, contaminants and high background can interfere with extraction of analytes at the MS level, even when using very high resolution. Using fragment ion data at the MS/MS level for peak extraction and integration can greatly improve the quality of the quantification, as the background and interferences can be virtually eliminated. Moreover, using MS/MS-level data relieves the burden of performing additional sample preparation steps or investigating longer, more protracted chromatographic gradients to clean up MS-level data.
As the sample complexity increases and/or faster chromatography is used, more compounds elute per given unit of time. This can further exacerbate any problems encountered at the MS level. Using high resolution at the MS/MS level allows high-quality extraction and integration of important fragment ions, even in the presence of interferences, enabling lower detection and quantitation limits and higher quantitative accuracy. Shorter gradient lengths also mean shorter cycle times are required to provide the minimum number of data points for every analyte. To provide good quantification, the cycle time should be fast enough so that each analyte is sampled a minimum of 10 times across its LC peak. The number of scans and accumulation times must therefore be carefully balanced to run at a cycle time which can deliver sufficient data points for the chosen chromatography. When it comes to MS/MS level data, DDA lacks the reproducibility and precision to generate the high-quality quantitative data required in biological studies.
The DIA (r)evolution
The main objective of DIA approaches is to fragment all analytes across the chosen m/z range regardless of their intensity. Thus, unlike DDA, the generation of MS/MS data is independent of precursor ion detection in a survey scan. However, interpretation of the resulting spectra can be challenging due to the complexity of the resulting MS/MS data. The power of any DIA-based analysis is therefore related to the level of selectivity prior to fragmentation and subsequent ion detection. The most selective of DIA methods is the infusion-based MS/MSALL workflow, which acquired product ion spectra of all precursors within a specified mass range. A typical experiment consisted of a TOF MS scan from m/z 200-1500 followed by a sequential acquisition of 1001 MS/MS spectra acquired from m/z 200.015 to 1200.051, with a step size of 1 Da. During the MS/MSALL experiment, a complete record of all precursors, product ions, and neutral losses within a sample was collected. This workflow is a powerful approach for discovery quantitation of lipids, but without orthogonal separation by chromatography this workflow was not applicable to proteomics analysis of complex samples.
In 2012, the data independent acquisition strategy known as SWATH-MS was published for the first time.5 With SWATH DIA, all ionizable precursors are analyzed by MS/MS regardless of abundance or other criteria. This results in greater reproducibility arising from a complete data set containing fragment data for all precursor ions. Although SWATH DIA was originally used for proteomics experiments, the workflow has been adopted for a wide variety of other applications, including metabolomics, environmental screening, food testing, forensics, and pharmaceutical analysis .6 With SWATH DIA, wider precursor selection windows are used for MS/MS that can allow multiple compounds through simultaneously. These windows are stepped across the entire precursor mass range such that all precursor masses are fragmented for every cycle. Compared with DDA, MS/MS spectra generated through SWATH DIA tend to be more complex. The use of variable window widths further increases the selectivity of SWATH DIA. With variable windows, the width of each precursor window is adjusted according to the density of the precursor ions present within that mass range. Very narrow windows are used where precursor ion density is greatest and wider windows are used where precursor ions are more sparsely populated. The increased selectivity and specificity can profoundly improve the quality of the resulting data. The precursor isolation window widths typically range from 2-50 Da (or wider). Even when two or more compounds fall within a SWATH DIA precursor isolation window, and have similar elution times, deconvolution of the MS/MS is usually possible by techniques based on LC profile correlation. However, when two or more compounds fall within the same precursor isolation window and have identical elution times, the deconvolution of the fragmentation signals is not possible. One solution was to design the SWATH DIA precursor isolation windows such that each internal standard had its own narrow isolation window. However, this made the method very specific to the compounds being measured. Adding new compounds to this method would require additional work, which negates one of the key benefits of SWATH acquisition, in that it is a generic and easy-to-set-up method. These issues can be compounded when chromatography is shortened to gain higher throughput. This would require using MS methods with lower overall acquisition times.
HT analytical methods are limited by the sampling rate of the MS instrument and the occurrence of signal interferences. Short duty cycle times are required to ensure that there are enough data points sampled for each chromatographic peak to provide accurate quantitation. To achieve this with fast gradients, DIA methods would have to sacrifice selectivity by using wider DIA isolation windows, or sensitivity by limiting the individual DIA window MS/MS scan times. Both factors can impact the accuracy of the quantitation and identification of peptides and proteins. These needs and challenges led to the development of an alternative DIA technique referred to previously as Scanning SWATH acquisition.7,8 Scanning SWATH acquisition continuously scans the quadrupole along the mass range, rather than in discrete steps as with SWATH DIA. Ion events are recorded along each TOF pulse in a synchronized fashion as Q1 is ramped. This way, every ion event is characterized by 3 independent coordinates: m/z, LC retention time, and the position of the Q1 window. The Q1 dimension provides information used to identify fragment ions by correlating their appearance/disappearance with the presence of a precursor in the Q1 isolation window at a given point in time. Additionally, this Q1 dimension from Scanning SWATH acquisition distinguishes precursor signal from any interfering signal at the same m/z that came from internal fragments, adducts or losses occurring at low collision energy, thereby resulting in cleaner spectra than with simple MS1 scans. However, the requirements to improve sensitivity and run faster remained.
Zeno trap technology
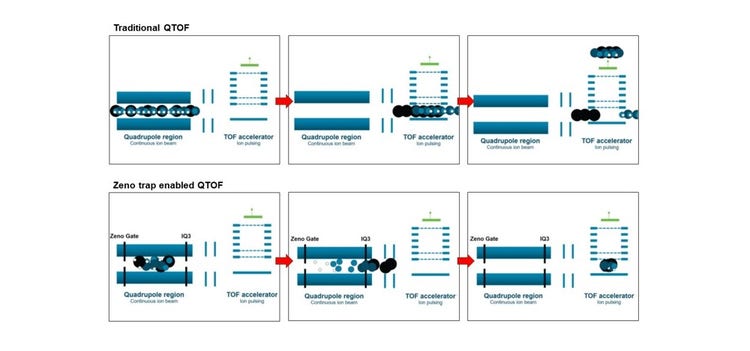
Quadrupole time-of flight (QTOF) instruments most commonly use the orthogonal injection of ions coming from a quadrupole collision cell into the flight tube region, because this configuration maximizes TOF resolution, mass accuracy, and sensitivity for an entire spectrum without the need for scanning. This type of ion pulsing, however, suffers from a relatively low duty cycle (Figure 3a). Typically, only 5-25% of ions are ejected with each pulse of the accelerator, depending on the geometry and m/z range. This is not typically needed in the MS1 dimension, since the ion currents generated by modern sources (such as the Turbo V ion source) and transmitted by modern ion capture technology (such as the QJet ion guide), need to be reduced to prevent saturation and to protect the longevity of TOF MS detectors. In the MS/MS dimension, however, an improvement in the duty cycle can lead to significant gains in sensitivity.
The ion losses are a result of the drift region between the collision cell and the TOF accelerator, in which ions are widely distributed positionally. Therefore, only a fraction of the fragment ion slice (residing in the proper location where pulsing is applied) are pulsed into the TOF flight tube and detected. A significant fraction of ions is lost with each pulse. Previously, there have been many attempts to overcome this lack of synchronicity. It has only been achieved, however, either for narrow mass ranges or at low acquisition frequency.
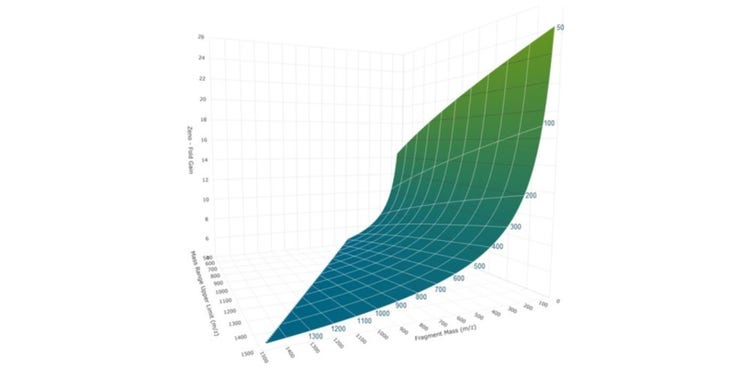
Use of the Zeno trap overcomes these technological barriers to recover duty cycle losses across the entire m/z at up to 133 Hz acquisition frequency (Figure 3b). This is achieved using a linear ion trap, referred to as a Zeno trap, at the exit of the collision cell. The mechanism of trapping and releasing ions is highlighted in Figure 3. Ions enter the ion trap and are contained with potential barriers on the ZG and IQ3 lenses, while subsequent packages of ions are accumulated in the LINAC collision cell, preventing ion loss. The trapped ions are left to energetically cool and are subsequently released based on potential energy resulting in an ordered release generally ranging from high m/z to low m/z. In this way, ions across the mass range reach the pulsing region of the TOF accelerator. This simple trapping and releasing mechanism leads to significant gains in MS/MS sensitivity, as highlighted in Figure 3. MS/MS with the Zeno trap activated results in a 4- to 15-fold (or greater) gain in signal, with increased gains at low m/z fragments. The Zeno trap efficiency combined with precise ion-release timing yields ≥90% of the theoretical gain across the entire mass range. Due to the degree of selectivity enabled with high resolution MS/MS data, these improvements in signal are combined with negligible changes to noise, resulting in spectral and chromatographic signal-to-noise on the order of the gains observed in raw signal (Figure 4).
Zeno SWATH DIA
With the incorporation of the Zeno trap to a SCIEX QTOF system, the SWATH DIA workflow is enhanced further. Zeno SWATH DIA combines the sensitivity enabled by the Zeno trap with the reproducibility and precision of SWATH DIA. The Zeno trap provides duty cycle improvements which boost sensitivity at MS2 level, delivering up to 3x more identified proteins, and ~3-6x more quantified, at loads less than 20 ng. High-quality MS/MS spectra are translated to the MS/MS extracted ion chromatograms (XICs) and to the total peptide ion current (TIC) permitting many identified proteins to be quantified reproducibly with <20% CV. 9
As MS technology continues to improve, high resolution instruments are more sensitive and capable of operating at faster scan rates, thereby pushing the boundaries of coverage depth at low loads while helping to address the demands of high-throughput OMICS workflows . Shorter gradient MS-based methods require lower cycle times to achieve results that strike a balance between the number of identifications and the precision of measurements. As LC throughput increases, conventional DIA faces the challenge of either decreasing MS/MS acquisition time, which reduces duty cycle, or increasing the window width, which reduces specificity. While Zeno SWATH DIA is indeed a powerful mechanism for protein identification and quantitation, it lacks the ability to assign precursor masses to the fragments in cases where the resulting MS/MS spectra are highly complex. In the case of shortest gradients, there are challenges to achieving specificity while maintaining necessary sampling points across the chromatographic peak for quantitation. This creates a need for a DIA mechanism that can run scans at a higher rate than Zeno SWATH DIA without compromising the specificity.
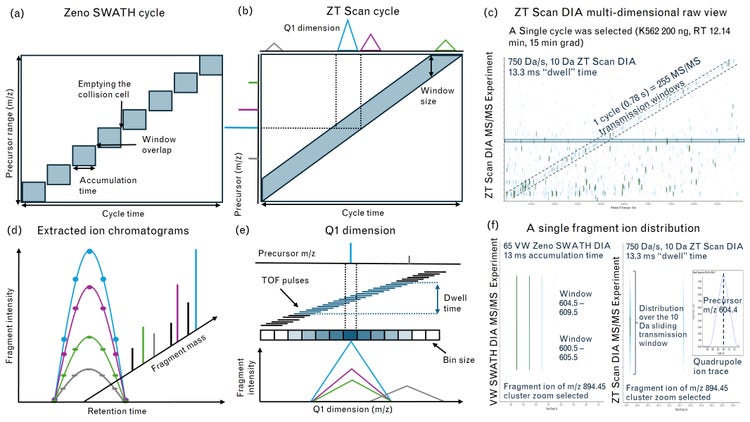
ZT Scan DIA
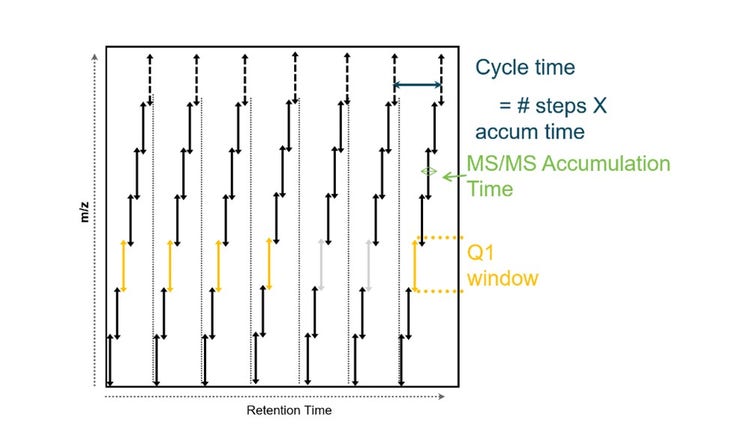
The scanning DIA methodology was implemented, on a research-modified Zeno trap enabled QTOF, which incorporates the sensitivity gains and duty cycle improvements enabled by Zeno trap. The ZT Scan DIA method offers two significant advantages over conventional DIA techniques. Zeno SWATH DIA requires the collision cell to be emptied between MS/MS acquisitions from the discrete DIA windows, leading to an overhead of 1-2 ms per MS/MS event (Figure 5a). As the acquisition speed increases, this overhead can reduce duty cycle, dropping below 80% at 6.6 ms per MS/MS. In contrast, the ZT Scan DIA approach does not have this limitation, allowing for higher duty cycles even at faster acquisition speeds. Secondly, in conventional DIA, fragment ions can only be assigned to precursors based on similar retention times within the width of the Q1 transmission window. However, ZT Scan DIA overcomes this limitation and enables more accurate precursor association. This reduces interferences from co-eluting isobaric analytes, contaminants, and high background noises, which impact the quantitation of analytes, especially at the MS level, even when using very high-resolution instruments. ZT Scan DIA maximizes the accuracy and precision of quantitation by utilizing the selectivity of MS/MS and the Q1 dimension. When combined with the highest scan rates and the use of a Zeno trap, mass resolution and accuracy are preserved to maintain the maximum number of identified and quantified analytes.
During each cycle, the isolation window for MS/MS is sliding along the m/z range of interest at an MS/MS acquisition rate. Based on the 400-900 m/z range method, scanning at 750 Da/s with a sliding isolation window of 5 Da, this would equate to a scan rate of 640 Hz. Ion events are recorded at each TOF pulse, as the isolation window scans across the mass range (Figure 5b). The scanning DIA mechanism does not require emptying the collision cell, thereby saving on cycle time. Each fragment is visible when the leading edge of the quadrupole transmits the precursor m/z and disappears when the trailing edge of the quadrupole passes it (Figure 5e). Thus, fragments can be mapped to the precursors bringing in an additional dimension along with the m/z, intensity, and RT dimensions (Figure 5d). This Q1 dimension from ZT scan DIA is used to distinguish precursor signal from any interfering signal at a different m/z that came from internal fragments, adducts or losses occurring at low collision energy (Figure 5e). Thus, results in improved deconvolution of complex analytes with a higher degree of specificity (Figure 5f).
This combination of fast scanning and sensitivity maximizes the total number of high-quality MS/MS spectra generated per cycle. This generates PRM level data for all ions in a sample, increasing specificity and, therefore, confidence in the total number of identified and quantified analytes. Faster scanning also enables the use of shorter LC run times, greatly improving throughput and laboratory productivity. This leads to a more comprehensive understanding of underlying biological changes. With ZT Scan DIA, maximal information is obtained from each precious sample. Consequently, each TOF experiment contains more useful MS/ MS information, particularly on lower abundance species that were previously undetectable, introducing researchers to a new level of sensitivity and specificity.
Conclusions
- Zeno trap activation brings significant sensitivity gains to MS/MS data acquisition and ZT scan DIA capitalizes on these gains by enabling the identification and quantification of significantly more analytes in a shorter time and with higher precision
- The additional selectivity provided by the quadrupole dimension further increases specificity to deliver unprecedented levels of analyte identification and quantification
- ZT Scan DIA provides both qualitative and quantitative data, combining the depth of coverage achieved with DIA and the specificity and precision of PRM workflows
References
- Chernushevich, I.V., Merenbloom, S.I., Liu, S., Bloomfield, N. A W-geometry ortho-TOF MS with high resolution and up to 100% duty cycle for MS/MS. J. Am. Soc. Mass Spectrom., 28, 2143-2150 (2017).
- Zeno trap: Defining new levels of sensitivity. (2021) SCIEX white paper, RUO-MKT-19-13373-B.
- Wang, Z. et al. High-throughput proteomics of nanogram-scale samples with Zeno SWATH DIA. BioRxiv preprint: https://www.biorxiv.org/content/10.1101/2022.04.14.488299v1.full
- Qualitative flexibility combined with quantitative power: Using the ZenoTOF 7600 system, powered by SCIEX OS software (2021) SCIEX technical note, RUO-MKT-02-13053-B.
- Gillet, L.C. et al. Targeted data extraction of the MS/MS spectra generated by data independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 (2012).
- SWATH® Acquisition Improves Metabolite Coverage over Traditional Data Dependent Techniques for Untargeted Metabolomics; RUO-MKT-02-7128A.
- Ivosev, G.; Cox, D.M.; Bloomfield, N. etc. Scanning SWATH Acquisition Method for Improved Compound Screening. 64th ASMS poster (2016).
- Messner, C.B; Demichev, V.; Bloomfield, N. etc. Ultra-fast Proteomics with Scanning SWATH. Nature Biotechnology. 39 (2021) 846-854.
- Collins, B.C., Hunter, C.L., Liu, Y. et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 8, 291 (2017). https://doi.org/10.1038/s41467-017-00249-5