Abstract
This technical note demonstrates the power and capability of the ZenoTOF 7600 hybrid time-of-flight mass spectrometry (TOFMS) system to sensitively detect, quantify, and structurally characterize the bile acid content of biological specimens. Analysis of bile acids by nominal mass instruments, such as triple-quadrupole MS (TQMS) systems, is challenging because of the high chemical background found in several precursor ions to precursor ion-based multiple-reaction monitoring (MRM) transitions used in current state-of-the-art assays (1-4). High-resolution mass spectrometry (HRMS) generates a full product ion spectrum for each targeted bile acid, and extracting fragment ions with a narrow mass-to-charge (m/z) window can reduce background chemical interferences and improve the signal-to-noise (S/N) of the assay. The detection of individual bile acid isomers currently depends on chromatographic resolution; collision-induced dissociation (CID)-based fragmentation cannot distinguish these isomeric metabolites. Electron-activated dissociation (EAD; 5-7) is a complementary fragmentation mode to CID that can provide structurally diagnostic fragment ions to differentiate bile acid isomers (Figure 1).
Herein, the ZenoTOF 7600 system was used to quantify the bile acid content of human plasma sample extracts with a sensitivity comparable to that of high-end TQMS systems. Furthermore, the EAD-based fragmentation generated several structurally diagnostic fragment ions that are capable of distinguishing between the bile acid isomers. These data demonstrate the capability of the ZenoTOF 7600 system to perform highly sensitive and selective analysis of endogenous bile acid content in human plasma.
Key features of bile acid analysis on the ZenoTOF 7600 system
- The ZenoTOF 7600 system can detect and quantify bile acids with a sensitivity comparable to that of high-end TQMS instruments
- The narrow fragment ion extraction window possible with HRMS improves assay sensitivity by reducing background chemical interferences and, consequently, increases the S/N of the assay
- EAD can generate diagnostic fragment ions to enable the distinction between bile acid isomers, which improves the overall specificity of the analysis
Introduction
Primary bile acids are cholesterol-derived molecules that are synthesized in the liver and are collected and pre-concentrated in the bile contained in the gall bladder (8). During gastric emptying, bile is secreted into the duodenum via the common bile duct, where bile acids play a crucial role in emulsifying and absorbing dietary fats (8,9). The two primary bile acids, cholic acid (CA) and chenodeoxycholic acid (CDCA) are bioconverted into chemically distinct, bacterially produced secondary bile acids and an expanding list of microbially-conjugated bile acids (MCBAs) by host microbiota (10-16). Of considerable recent interest is the potential role of these bacterial-derived secondary bile acids in human physiology and their contributions to pathologies such as inflammatory bowel disease and cancer. Consequently, there is great interest in identifying and quantifying these compounds in diverse samples.
Multiple methods for bile acid analysis have been reported in the literature (1-4). These methods are typically TQMS-based, and their effectiveness highly depends on sample prep, instrument sensitivity, and chromatography to deliver high-quality bioanalytical results. One common drawback of these published methods is that they often use the negative ion mode. At the mass range of interest (~400-500 Da), there is considerable isobaric chemical interference from the matrix and the mobile phase solvents, depending on their source. This is especially problematic for low-resolution TQMS instruments (Rs ~100-150 equating to ±0.1 Da mass error). HRMS offers a potential solution to this problem due to its high mass accuracy and high resolution. Fragment ions can be extracted from a high-resolution MS/MS spectrum using a narrow m/z tolerance window (~ 20 mDa), which may exclude many isobaric interferences that cause the high background in TQMS instruments. The ZenoTOF 7600 system can acquire data in this mass range with a mass accuracy of ± 2ppm and a resolution of >35K.
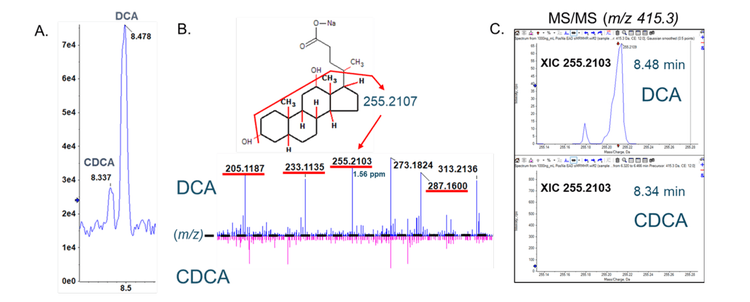
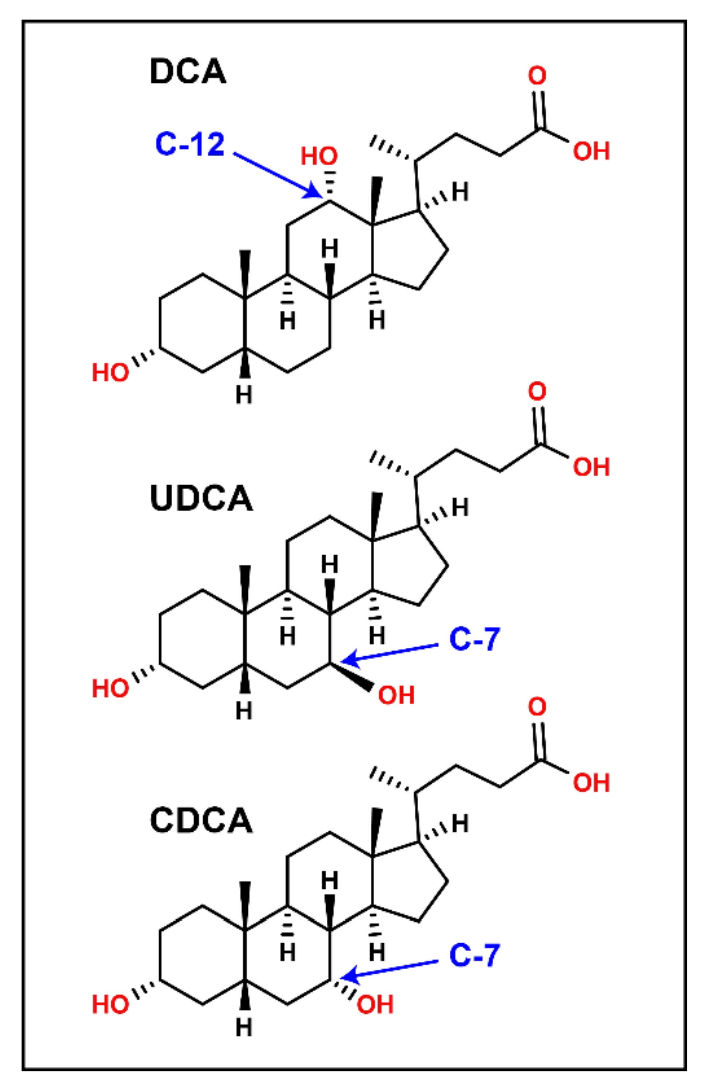
Bile acids are cholesterol-derived amphipathic molecules of saturated hydroxylated C-24 sterols. As shown in Figure 2, the location and stereochemistry of the hydroxyl functional groups define the different isomers within each sub-class. CID-based fragmentation of bile acids primarily yields product ions related to the bile acid head group, and relatively few fragments are generated from the ring structure; hence, CID alone does not generate diagnostically useful fragment ions for identification or quantification purposes. Using chromatography to resolve bile acid isomers requires relatively long chromatographic gradients, which are not conducive to high-throughput analysis. An alternative to lengthy chromatographic gradients and CID-based fragmentation is EAD. EAD-based fragmentation provides ample fragments derived from the sterol ring structure (Figure 1A & 1B), and it can distinguish the isomers of each bile acid subclass without extensive chromatographic method development.
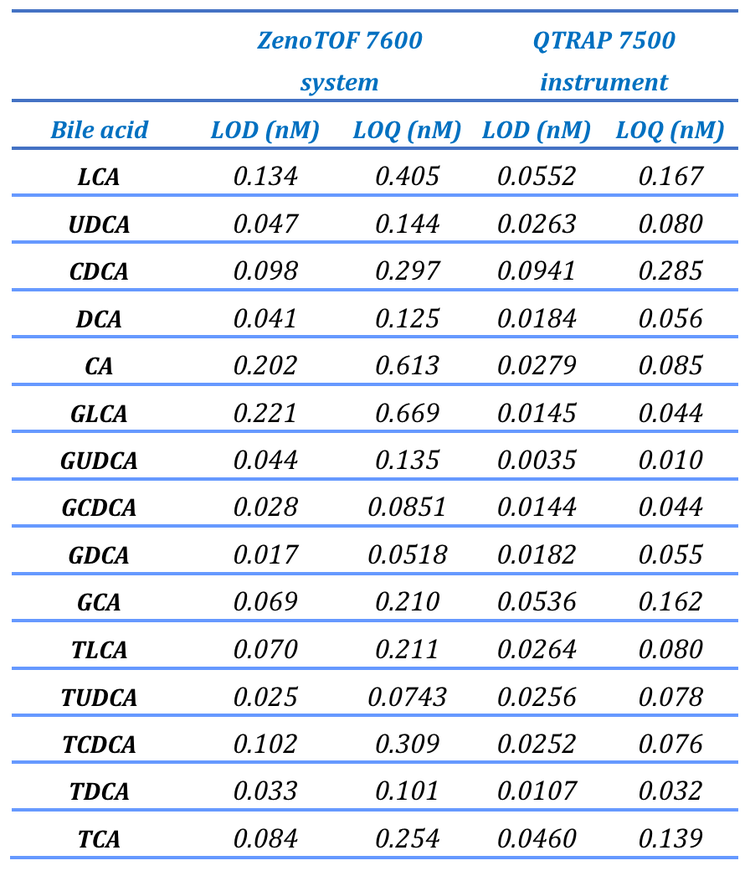
The ZenoTOF 7600 system has an EAD cell that provides complimentary fragmentation that can be activated and scheduled within a typical quantitative bioanalytical method to provide structural characterization of bile acids. In this technical note, the speed and sensitivity of the ZenoTOF 7600 system were leveraged to quantify bile acids in prepared plasma samples. These results were compared to those acquired on the QTRAP 7500 instrument. To demonstrate the power of EAD for structural characterization, the DCA sub-class was structurally characterized to identify EAD-specific fragments for each isomer.
Methods
Materials: Bile acid standard mixtures were purchased from Cambridge Isotopes Laboratories. The stable isotope-labeled bile acid mixes (unconjugated, cat# MSK-BA1; conjugated, MSK-BA2) were used as internal standards, and unlabeled bile acid mixes (unconjugated, cat# MSK-BA1-US; conjugated, MSK-BA2-US) were used as primary reference standards. All solvents were LCMS grade and obtained from Burdick and Jackson and Fisher Scientific.
Sample preparation: Human blood (research use only) was collected into tubes containing potassium ethylenediamine tetraacetic acid (K2EDTA) anti-coagulant following the procedure outlined in the institutional review board (IRB) protocol #______. The plasma was isolated from the red blood cell fraction by centrifugation. A 50 µL volume of each plasma sample was pipetted into individual 0.6 mL Eppendorf tubes, and a 200 µL volume of methanol was added to precipitate plasma proteins (primary DF=5-fold). All samples were vortex-mixed for 30 seconds and centrifuged at 17,000g for 5 minutes, and the supernatant for each clarified plasma extract sample was transferred into fresh vials.
A 50 µL volume of each clarified plasma extract sample was transferred into fresh glass vials. A 450 µL volume of an internal standard solution (10 nM deuterated BAs) was added to each sample vial (DF=50-fold overall), and the vials were capped and vortex-mixed for 30 seconds. The final volume of each sample was split into two identically labeled glass autosampler vials (w/ inserts), with each set being injected on the QTRAP 7500 and the ZenoTOF 7600, respectively.
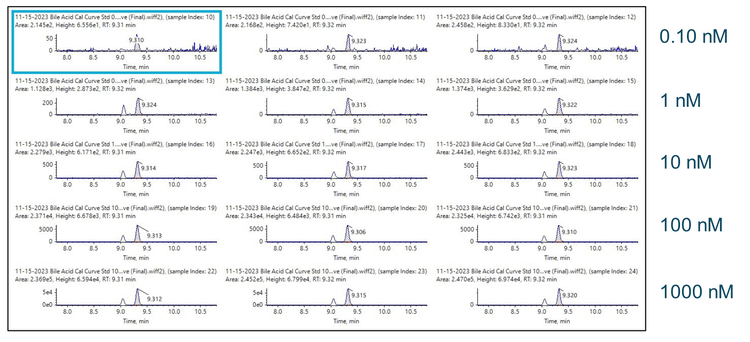
Chromatography: Extracted metabolites were resolved using two different HPLC systems. For the ZenoTOF 7600-based system, an Exion UHPLC instrument equipped with a Phenomenex Kinetex XBC18 column (100 x 2.1 mm; 2.6 µm particle size) was used. For the QTRAP 7500 TQMS-based system, a Nexera 40 Series UHPLC system equipped with a Restek Raptor C18 column (100 x 2.1 mm; 5.0 µm particle size) and an Ultra C18 guard column (10 x 2.1 mm i.d., 5 µm; Restek) was used. The two different columns produced similar but not identical chromatographic elution profiles. Notably, both columns entirely resolved the multiple bile acid isomers. The autosampler sample bay was maintained at 10 °C for both systems, and a 15 µL sample volume was injected on column. The column oven temperature was kept at 50oC with a constant mobile phase flow rate of 0.3 mL/min for a total run time of 17 min. The mobile phase compositions were (A) 10 mM ammonium formate in water and (B) pure acetonitrile. Gradient details are shown in Table 1
Mass spectrometry: Extracted sample extracts were analyzed using the ZenoTOF 7600 system with an OptiFlow Turbo V ion source and a QTrap 7500 system with an OptiFlow Pro Turbo V ion source using a scheduled, high-resolution multiple reaction monitoring (sMRMHR) or a scheduled multiple reaction monitoring (sMRM) scan mode, respectively. In some cases, a pseudo-MRM transition was used (i.e., precursor ion to precursor ion). On the QTRAP 7500 system, these molecules were quantitated at the MS/MS level with low CE. However, for the data acquired for these bile acids on the ZenoTOF 7600 system, the TOFMS data were used.
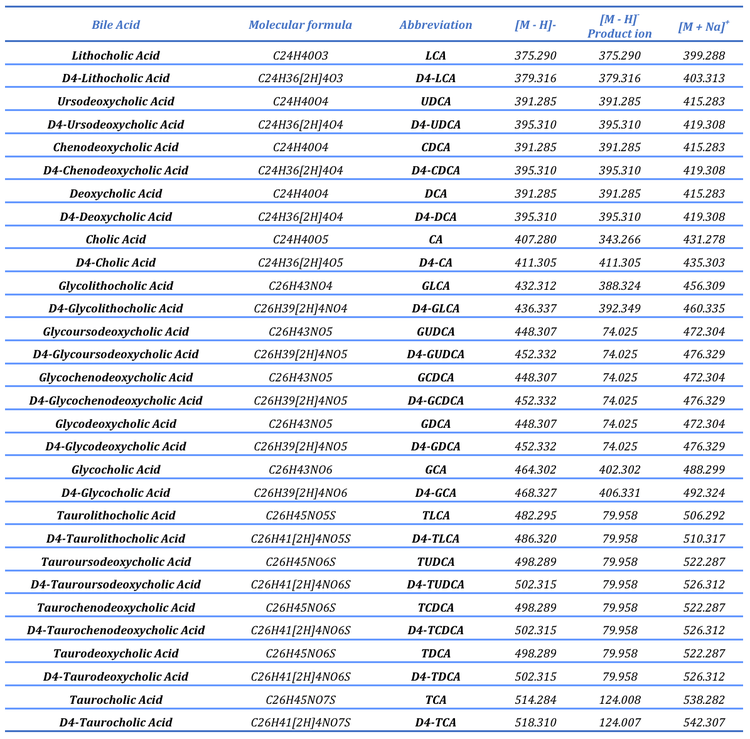
To structurally characterize bile acids, an MRM-triggered datadependent acquisition (DDA) scan mode was used to acquire EADbased product ion spectra in the positive ion mode (standards only). Although EAD-based fragmentation is possible in the negative ion mode, it is relatively inefficient compared to the positive ion mode; consequently, the EAD-based analysis was performed in the positive ion mode. Because bile acids are anionic, the sodium adduct for each was was monitored and isolated for fragmentation. A summary of the MS instrument parameters is presented in Tables 2 and 3, and a comprehensive list of the compounds studied is shown in Table 4.
Results and Discussion
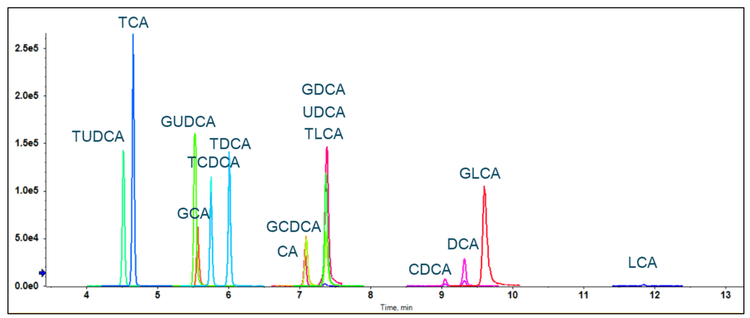
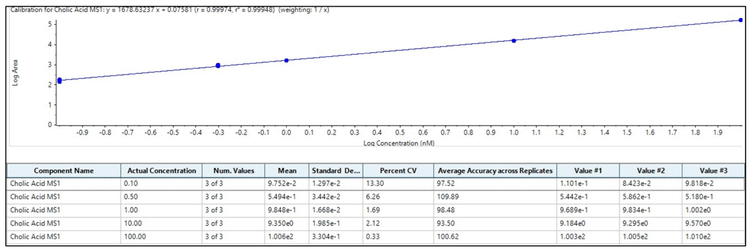
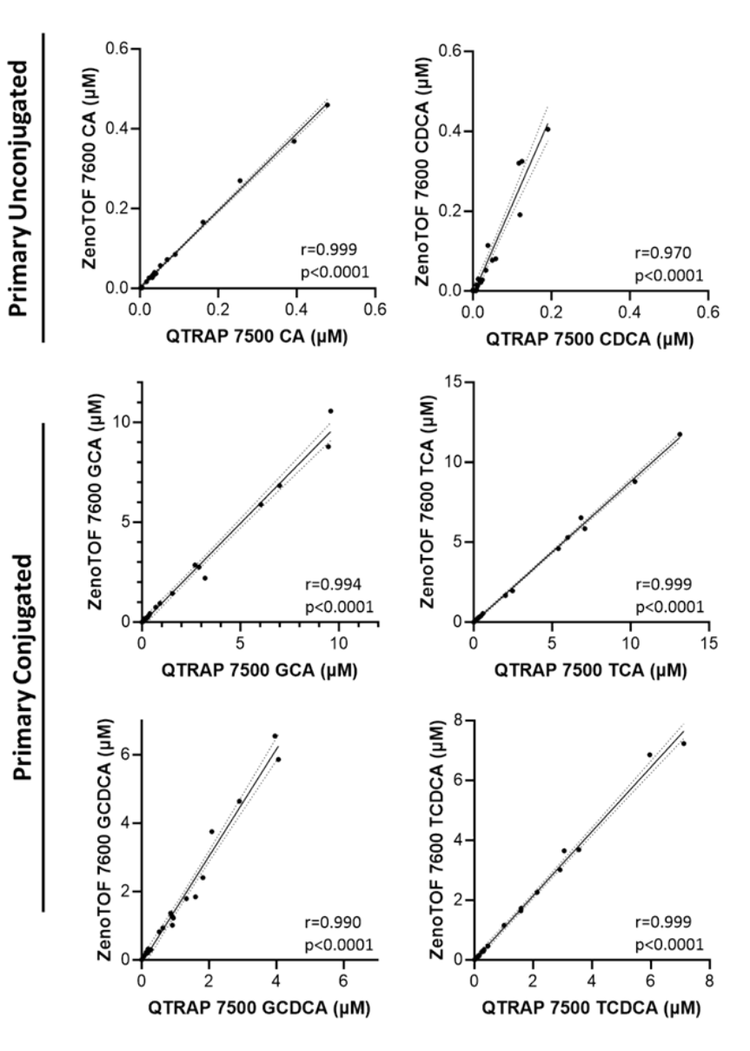
A bile acid standard curve was generated neat in solvent and analyzed on the QTRAP 7500 and the ZenoTOF 7600 systems. The parameter settings were optimized for each instrument, as detailed in Tables 2 and 3, and the chromatographic gradient conditions resulted in the separation of each isomeric bile acid with baseline resolution (Figure 3). The concentrations within the internal standard curve ranged from 0.01 to 1000 nM, covering a dynamic range > 5 orders of magnitude. Due to the sensitivity of the QTRAP 7500 instrument, the 1000 nM concentration standards caused detector saturation; this was not an issue on the ZenoTOF 7600 system. Consequently, to better match the conditions on each instrument, the final standard curve calculations and subsequent in vivo measurements were truncated to a linear dynamic range of 0.1 to 100 nM. Due to this consideration, it is important to note that the results presented here do not necessarily reflect the maximum extent of the dynamic range capability of the instrument for the measurement of bile acids. The range tested, however, was sufficient to measure bile acids in rat plasma. An example curve acquired for cholic acid on the ZenoTOF 7600 system and its associated ion statistics are presented in Figure 4.
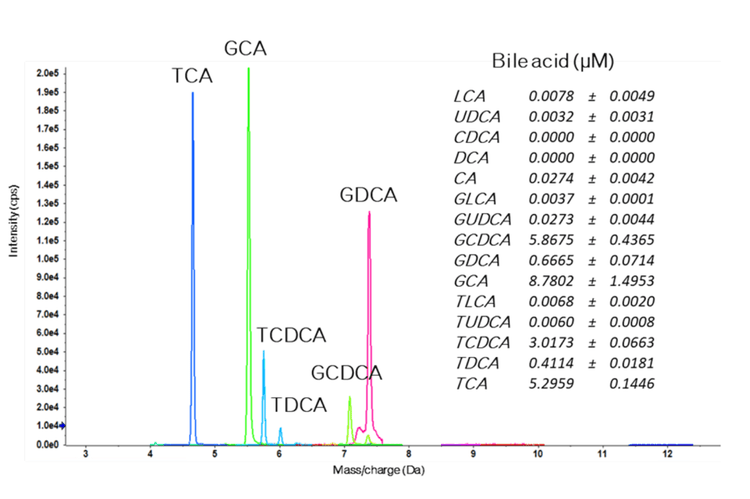
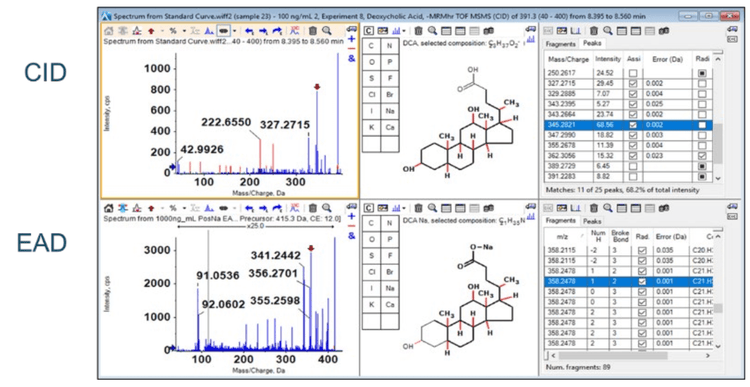
The EAD-based fragmentation functionality of the ZenoTOF 7600 system can provide significantly more structural details than that generated by CID-based fragmentation. Figure 9 shows the product ion spectrum for DCA. This experiment analyzed a DCA standard (100 nM) using the ZenoTOF 7600 system by CID- and EAD-based fragmentation. The CID spectrum in the top left panel has relatively few fragments compared to the EAD-based spectrum in the bottom left panel. The middle panels show the .mol files for DCA and indicate the highlighted fragment in the right panels in bold. This feature of SCIEX OS, called fragment pane, enables the structural characterization of compounds by matching the actual MS/MS data to in silico-generated fragments based on the .mol file. The predicted CID fragments are shown in red in the top left panel. Note that most of these fragments are not present in the spectrum. However, all the predicted fragments appear for DCA fragmented via EAD. This ability to generate fine structural detail was extended to determine whether EAD-based fragmentation could distinguish bile acid isomers.
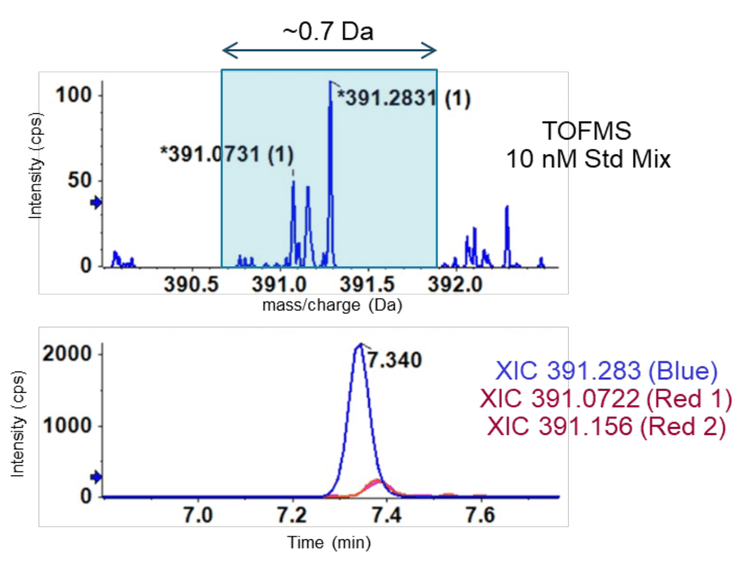
Figure 2 shows that DCA, CDCA, and UDCA have the same empirical formula and exact mass but differ structurally in the position and stereochemistry of their hydroxyl functional groups. Figure 1A shows the TOFMS total ion chromatogram (TIC) for m/z 415.3 (DCA sodium adduct) and the importance of chromatography in resolving bile acid isomers. Using EAD-based fragmentation, unique fragment ions were identified for DCA and CDCA, the two isomers that elute closely (Figure 1B). These fragments are underlined in red on the spectra. Notably, the DCA-specific fragment at m/z 255.2107 shows no interference in the XIC for chromatographically resolved CDCA. Thus, the m/z fragment at 255.2017 Da can be used as a DCAspecific fragment for sMRMHR analysis and may enable a shorter gradient for higher throughput. EAD-based fragmentation may enable the complete structural characterization of these molecules without requiring purification or other structural diagnostic tools. More importantly, this type of fragmentation may help identify novel conjugated bile acids (10-15). For example, Quinn et al. recently reported the discovery of several new MCBAs (specifically phenylalanocholic acid (Phe-CA), tyrosocholic acid (Tyr-CA), and leucocholic acid (Leu-CA)) that were discovered in the intestinal contents collected from mice (11). Another study found evidence of Figure 9. Product ion analysis of deoxycholic acid (DCA). A DCA standard (100 nM) was analyzed using the ZenoTOF 7600 system by CID- and EAD-based fragmentation (left panels). The middle panels show the .mol file for DCA, and the panels on the right show the theoretical fragments for CID (i.e., heterolytic fragmentation) and EAD (radical-based fragmentation). The highlighted fragment in blue corresponds to structure highlighted in bold on the middle figures and is indicated by a red arrow in the MS/MS spectra on the left. 9 the amino acids glutamate, glutamine, aspartate, asparagine, methionine, histidine, lysine, serine, tryptophan, valine, alanine, and arginine forming conjugates with CDCA, DCA, or CA (12). Considering the emerging importance of MCBAs in human disease (19-20) and the critical role of mass spectrometry in their characterization (21), the quantitative sensitivity and qualitative capabilities of the ZenoTOF 7600 may prove invaluable to the growth of bile acid metabolism research.
Conclusion
- The specific method details are provided for the analysis of endogenous bile acids using the ZenoTOF 7600 system and the QTRAP 7500 instrument
- The ZenoTOF 7600 system is capable of measuring endogenous bile acids with comparable sensitivity to the QTRAP 7500 system
- The narrow XIC window possible on HRMS systems enables the MS1-based collection of data without the inclusion of chemical background interferences
- EAD-based fragmentation provides structural characterization of molecules and may abrogate the need for extensive chromatographic separation of isomers to ensure specificity
References
- Liu D, Li G, Liu D, Shi W, Wang H, Zhang Q, Shen M, Huang X, Lin H. Quantitative Detection of 15 Serum Bile Acid Metabolic Products by LC/MS/MS in the Diagnosis of Primary Biliary Cholangitis. Chem Biodivers. 2023; Mar;20(3):e202200720. doi: 10.1002/cbdv.202200720
- Gao T, Hu S, Xu W, Wang Z, Guo T, Chen F, Ma Y, Zhu L, Chen F, Wang X, Zhou J, Lv Z, Lu L. Targeted LC‐MS/MS proϔiling of bile acids reveals primary/secondary bile acid ratio as a novel biomarker for necrotizing enterocolitis. Anal Bioanal Chem. 2024; Jan;416(1):287-297. doi: 10.1007/s00216-023-05017-7
- Yang J, Lin J, Wang A, Yang X, Wang Y, Zhang Y, Dong H, Tian Y, Zhang Z, Wang M, Song R. Study on the effect of calibration standards prepared with different matrices on the accuracy of bile acid quantiϔication using LC‐MS/MS. J Pharm Biomed Anal. 2024; 237:115785. doi: 10.1016/j.jpba.2023.115785.
- Engevik MA, Thapa S, Lillie IM, Yacyshyn MB, Yacyshyn B, Percy AJ, Chace D, Horvath TD. Repurposing dried blood spot device technology to examine bile acid proϔiles in human dried fecal spot samples. Am J Physiol Gastrointest Liver Physiol. 2024; 326:G95–G106. Doi: 10.1152/ajpgi.00188.2023
- Baba T, Campbell JL, Le Blanc JCY, Baker PRS, Hager JW, Thomson BA. Development of a Branched Radio‐Frequency Ion Trap for Electron Based Dissociation and Related Applications. Mass Spectrom (Tokyo). 2017;6(1):A0058. doi: 10.5702/massspectrometry.A0058.
- Che P, Davidson JT, Kool J, Kohler I. Electron activated dissociation ‐ a complementary fragmentation technique to collision‐induced dissociation for metabolite identiϔication of synthetic cathinone positional isomers. Anal Chim Acta. 2023; 1283:341962. doi: 10.1016/j.aca.2023.341962
- Baba T, Campbell JL, Le Blanc JCY, Baker PRS. Distinguishing Cis and Trans Isomers in Intact Complex Lipids Using Electron Impact Excitation of Ions from Organics Mass Spectrometry. Anal Chem. 2017 Jul 18;89(14):7307-7315. doi: 10.1021/acs.analchem.6b04734. Epub 2017 Jun 28. PMID: 28613874.
- Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003; 72:137-74. doi: 10.1146/annurev.biochem.72.121801.161712
- Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006 ;47(2):241-59. doi: 10.1194/jlr.R500013-JLR200
- Guzior DV, Quinn RA. Review: microbial transformations of human bile acids. Microbiome. 2021; 9(1):140. doi: 10.1186/s40168-021-01101-1
- Quinn RA, Melnik AV, Vrbanac A, Fu T, Patras KA, Christy MP, Bodai Z, Belda-Ferre P, Tripathi A, Chung LK, Downes M, Welch RD, Quinn M, Humphrey G, Panitchpakdi M, Weldon KC, Aksenov A, da Silva R, Avila-Pacheco J, Clish C, Bae S, Mallick H, Franzosa EA, Lloyd-Price J, Bussell R, Thron T, Nelson AT, Wang M, Leszczynski E, Vargas F, Gauglitz JM, Meehan MJ, Gentry E, Arthur TD, Komor AC, Poulsen O, Boland BS, Chang JT, Sandborn WJ, Lim M, Garg N, Lumeng JC, Xavier RJ, Kazmierczak BI, Jain R, Egan M, Rhee KE, Ferguson D, Raffatellu M, Vlamakis H, Haddad GG, Siegel D, Huttenhower C, Mazmanian SK, Evans RM, Nizet V, Knight R, Dorrestein PC. Global chemical effects of the microbiome include new bile‐acid conjugations. Nature. 2020; 579(7797):123-129. doi: 10.1038/s41586-020-2047-9
- Lucas LN, Barrett K, Kerby RL, Zhang Q, Cattaneo LE, Stevenson D, Rey FE, Amador-Noguez D. Dominant Bacterial Phyla from the Human Gut Show Widespread Ability To Transform and Conjugate Bile Acids. mSystems. 2021; e0080521. doi: 10.1128/mSystems.00805-21
- Foley MH, Walker ME, Stewart AK, O'Flaherty S, Gentry EC, Patel S, Beaty VV, Allen G, Pan M, Simpson JB, Perkins C, Vanhoy ME, Dougherty MK, McGill SK, Gulati AS, Dorrestein PC, Baker ES, Redinbo MR, Barrangou R, Theriot CM.Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difϔicile growth in the murine gut. Nat Microbiol. 2023; 8(4):611-628. doi: 10.1038/s41564-023-01337-7
- Gentry EC, Collins SL, Panitchpakdi M, Belda-Ferre P, Stewart AK, Carrillo Terrazas M, Lu HH, Zuffa S, Yan T, Avila-Pacheco J, Plichta DR, Aron AT, Wang M, Jarmusch AK, Hao F, SyrkinNikolau M, Vlamakis H, Ananthakrishnan AN, Boland BS, Hemperly A, Vande Casteele N, Gonzalez FJ, Clish CB, Xavier RJ, Chu H, Baker ES, Patterson AD, Knight R, Siegel D, Dorrestein PC. Reverse metabolomics for the discovery of chemical structures from humans. Nature. 2024; 626(7998):419-426. doi: 10.1038/s41586-023-06906-8
- Guzior DV, Okros M, Shivel M, Armwald B, Bridges C, Fu Y, Martin C, Schilmiller AL, Miller WM, Ziegler KM, Sims MD, Maddens ME, Graham SF, Hausinger RP, Quinn RA. Bile salt hydrolase acyltransferase activity expands bile acid diversity. Nature. 2024; Online Ahead of Print. doi: 10.1038/s41586-024- 07017-8.
- Horvath TD, Haidacher SJ, Hoch KM, Auchtung JM, Haag AM. A high‐throughput LC‐MS/MS method for the measurement of the bile acid/salt content in microbiome‐derived sample sets. MethodsX. 2020; 7:100951. doi: 10.1016/j.mex.2020.100951
- ICH harmonized tripartite guideline: validation of analytical procedures: text and methodology Q2(R1), International Conference of harmonization of technical requirements for registration of pharmaceuticals for human use, 2005
- Baker, PRS and Proos R. Untargeted data-dependent acquisition (DDA) metabolomics analysis using the ZenoTOF 7600 system. SCIEX technical note, RUO MKT-02-15367-A
- McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y, DeRight Goldasich L, Dorrestein PC, Dunn RR, Fahimipour AK, Gaffney J, Gilbert JA, Gogul G, Green JL, Hugenholtz P, Humphrey G, Huttenhower C, Jackson MA, Janssen S, Jeste DV, Jiang L, Kelley ST, Knights D, Kosciolek T, Ladau J, Leach J, Marotz C, Meleshko D, Melnik AV, Metcalf JL, Mohimani H, Montassier E, Navas-Molina J, Nguyen TT, Peddada S, Pevzner P, Pollard KS, Rahnavard G, Robbins-Pianka A, Sangwan N, Shorenstein J, Smarr L, Song SJ, Spector T, Swafford AD, Thackray VG, Thompson LR, Tripathi A, Vázquez-Baeza Y, Vrbanac A, Wischmeyer P, Wolfe E, Zhu Q; The American Gut Consortium; Knight R. American Gut: an Open Platform for Citizen Science Microbiome Research. mSystems. 2018; 3(3):e00031-18. doi: 10.1128/mSystems.00031-18
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, AvilaPacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA 3rd; IBDMDB Investigators; Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C. Multi‐omics of the gut microbial ecosystem in inϔlammatory bowel diseases. Nature. 2019; 569(7758):655-662. doi: 10.1038/s41586-019-1237-9
- Wang M, Jarmusch AK, Vargas F, Aksenov AA, Gauglitz JM, Weldon K, Petras D, da Silva R, Quinn R, Melnik AV, van der Hooft JJJ, Caraballo-Rodrı́guez AM, Nothias LF, Aceves CM, Panitchpakdi M, Brown E, Di Ottavio F, Sikora N, Elijah EO, Labarta-Bajo L, Gentry EC, Shalapour S, Kyle KE, Puckett SP, Watrous JD, Carpenter CS, Bouslimani A, Ernst M, Swafford AD, Zúñiga EI, Balunas MJ, Klassen JL, Loomba R, Knight R, Bandeira N, Dorrestein PC. Mass spectrometry searches using MASST. Nat Biotechnol. 2020; 38(1):23-26. doi: 10.1038/s41587-019-0375- 9



