Abstract
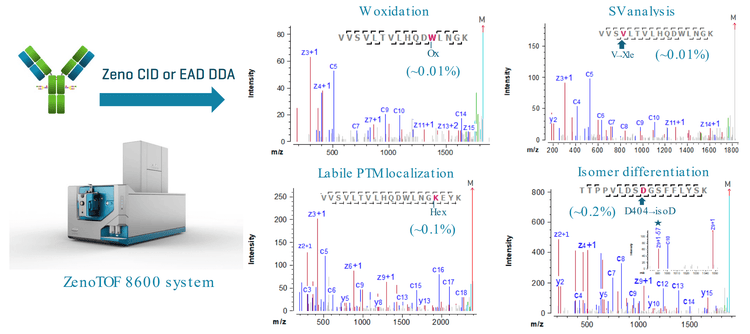
Comprehensive characterization of biotherapeutic product quality attributes (PQAs) requires high MS sensitivity for PQA detection and the ability of MS/MS to identify and localize labile post-translational modifications (PTMs), perform disulfide bond mapping and differentiate amino acid isomers.1-8 This technical note demonstrates an enhanced peptide mapping workflow with CID or electron-activated dissociation (EAD) for sensitive detection and confident characterization of low-abundant PQAs in biotherapeutics. The significant MS sensitivity gain offered by the novel ZenoTOF 8600 system with enhanced hardware components, such as the DJet ion guide, QJet ion guide, OptiFlow Pro ion source and TOF optics, enables routine analysis of PQAs below the trace level (<0.1%, Figure 1). This analytical advancement expands the capabilities of EAD for routine to advanced PQA characterization, facilitating biotherapeutic development and important decision-making
throughout the pipeline.
Key features of the enhanced peptide mapping workflow for biotherapeutic characterization
- Sensitive detection and confident identification of low-abundant PQAs: The improved MS sensitivity leads to comprehensive characterization of challenging PQAs down to the trace level (<0.1%)
- High sequence coverage: CID or EAD DDA provides nearly complete sequence coverage in a single injection with reduced sample load
- Localization of low-abundant labile PTMs: EAD DDA with improved MS sensitivity enables sensitive detection and accurate localization of labile PTMs, such as Man5 (~0.2%)
- Confident isomer differentiation: EAD generates signature fragments for clear differentiation of amino acid isomers
- Disulfide bond mapping: CID and EAD provide complementary results for confident disulfide bond mapping
Introduction
Biotherapeutics are highly complex and heterogeneous molecules containing various low-abundant PQAs, such as PTMs, amino acid isomers and sequence variants (SVs). The identification, characterization and monitoring of PQAs are essential components of biotherapeutic development to ensure the safety, efficacy and quality of drug molecules.1
Previous studies have demonstrated the advantages of EAD over traditional electron- or collision-based MS/MS approaches for comprehensive biotherapeutic characterization.2-8 EAD provides superior fragmentation spectra to allow accurate localization of labile post-translational modifications (PTMs) and clear differentiation of amino acid isomers. One challenge with PQA analysis using electron-based MS/MS approaches is to obtain sufficient signal of fragments for detailed characterization of low-level PQAs. This challenge was previously addressed using the Zeno trap, which increases the detection of MS/MS fragments by 5-10 fold. The challenge was further overcome by up to 10X gain in MS sensitivity provided by the novel ZenoTOF 8600 system with enhanced hardware components, such as the DJet ion guide, QJet ion guide,
OptiFlow Pro ion source, and TOF optics.
Methods
Sample preparation: The reduced NISTmAb (RM 8671, NIST) sample was prepared by denaturing the mAb with guanidine hydrochloride, followed by reduction using dithiothreitol (DTT) and alkylation with iodoacetamide (IAM). The non-reduced sample was prepared without using DTT. Enzymatic digestion of the reduced or non-reduced NISTmAb was performed using the trypsin/Lys-C protease (Promega). 100 ng-1 µg of the final digest was injected for LC-MS/MS analyses.
Liquid chromatography: Tryptic peptides were separated with the gradient displayed in Table 1 using an ACQUITY BEH C18 column (2.1 × 150 mm, 1.7 µm, 130 Å, Waters). A flow rate of 0.25 mL/min was used for the chromatographic separation. The column was kept at 60°C in the column oven of an ExionLC AD system (SCIEX). Mobile phase A was 0.1% formic acid (FA) in water and mobile phase B was 0.1% FA in acetonitrile.
Mass spectrometry: The peptide mapping data were acquired using a data-dependent acquisition (DDA) method with CID or EAD on a ZenoTOF 7600 system or ZenoTOF 8600 system. Table 2 shows key MS/MS parameters of CID or EAD DDA experiments conducted on the ZenoTOF 8600 system.
Enhanced sequence analysis using DDA methods with improved MS sensitivity
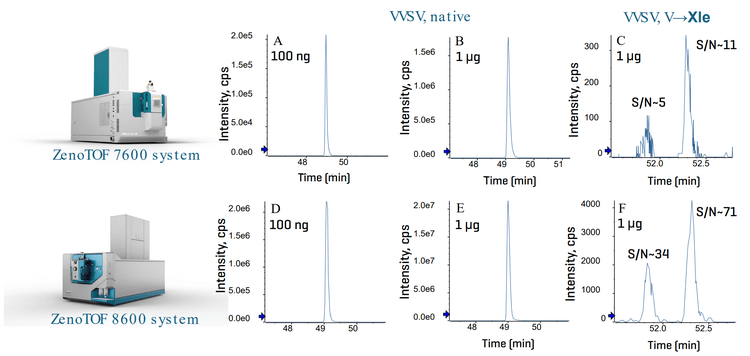
The improvement in the ion source, optics and detector of the ZenoTOF 8600 system results in up to 10X gain in MS sensitivity compared to the ZenoTOF 7600 system, providing significant benefits for the sensitive detection and confident identification of low-level PQAs or impurities in biotherapeutics. Figure 2 shows the extracted ion chromatograms (XICs) of the native VVSVLTVLHQDWLNGK (VVSV) peptide and its V→Xle (Xle = Ile or Leu) variants obtained from 100 ng or 1 µg injection of NISTmAb digest using the ZenoTOF 7600 system or ZenoTOF 8600
system. Compared to the data from the ZenoTOF 7600 system (Figures 2A-2C), the ZenoTOF 8600 system provided up to 10X increase in S/N with the same sample load or similar signal with 10X reduction in sample consumption (Figures 2D-2F). The MS sensitivity improvement leads to enhanced performance of the peptide mapping workflow using CID or EAD DDA.
Table 3 shows the peptide mapping results of 100 ng or 1 µg NISTmAb obtained using CID or EAD DDA. The enhanced peptide mapping workflow provided an improved sequence coverage of heavy chain (HC) and light chain (LC), particularly for EAD with the lower sample load (100 ng). A nearly complete sequence coverage (94-99%) of NISTmAb was achieved for a 100 ng sample load using CID or EAD DDA on the ZenoTOF 8600 system. By comparison, <90% sequence coverage was obtained using EAD DDA on the previous system. Additionally, the enhanced peptide mapping workflow provided a significant increase (~2X) in the number of identified peptides (ID #), including many in low abundance, increasing the depth of biotherapeutic characterization. Selected examples of low-abundant PQAs that are challenging to characterize using the traditional peptide mapping workflow on the ZenoTOF 7600 system will be presented in the following sections.
Sensitive detection and confident identification of low-abundant PTMs
The increasing complexity of new-generation biotherapeutics calls for advanced analytical techniques that can achieve straightforward and comprehensive PQA characterization with high sensitivity and broad capabilities.1 The EAD-based peptide mapping workflow has proven to be highly effective for sequence confirmation, PTM localization, disulfide bond mapping and isomer differentiation.2-8 The gain in MS sensitivity provided by the ZenoTOF 8600 system further enhances the capabilities of EAD for in-depth characterization of low-abundant PTMs or impurities in biotherapeutics.
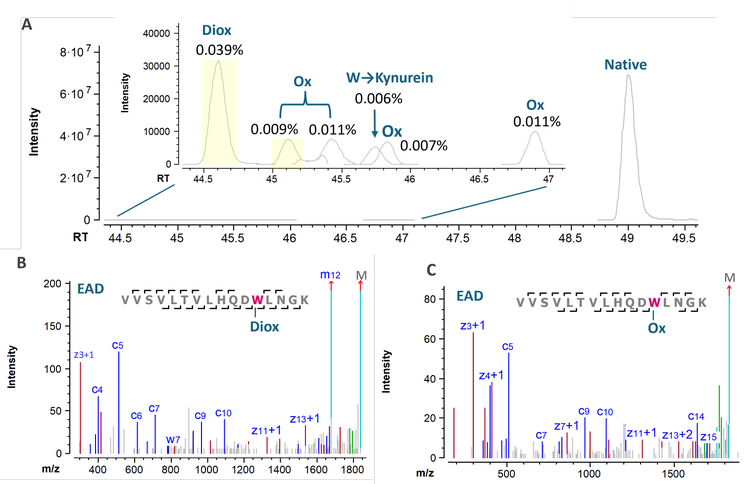
Figure 3 shows the XICs and selected EAD spectra of tryptophan (W) oxidation species of the VVSV peptide. These species are challenging to detect and identify using DDA on the previous platform. The MS sensitivity increase offered by the ZenoTOF 8600 system enabled sensitive detection of W oxidation species as low as 0.006% from DDA runs (Figure 4A). Benefiting from the MS improvement, EAD generated extensive
fragments of these low-abundant species for confident sequence identification and accurate localization of the modification.
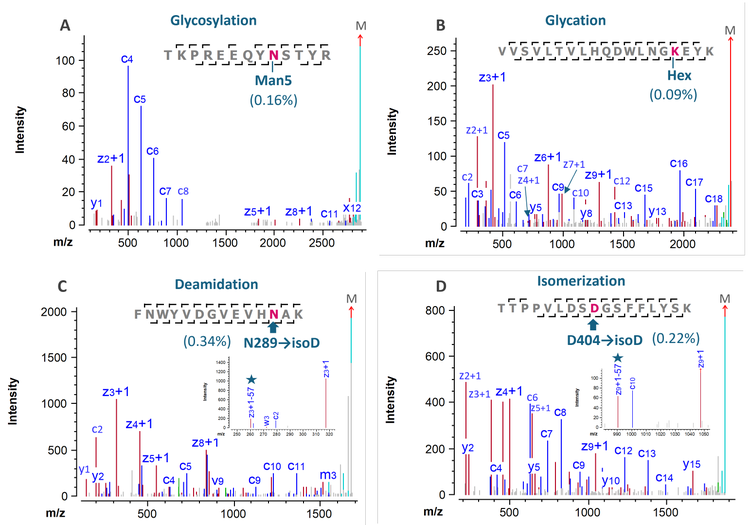
The enhanced peptide mapping workflow using EAD provided comprehensive characterization of labile PTMs in low abundance, such as a glycopeptide carrying Man5 (0.16%, Figure 4A) and a glycated species of the VVSV peptide (~0.1%, Figure 4B). The ability of EAD to preserve labile PTMs enabled precise localization of glycosylation and glycation (Hex) in the respective peptide. For example, the detection of fragments z2 and z3 without Hex and z4 with Hex pinpointed the position of glycation in the glycated VVSV peptide (Figure 4B).
Enhanced Sequence Variant analysis
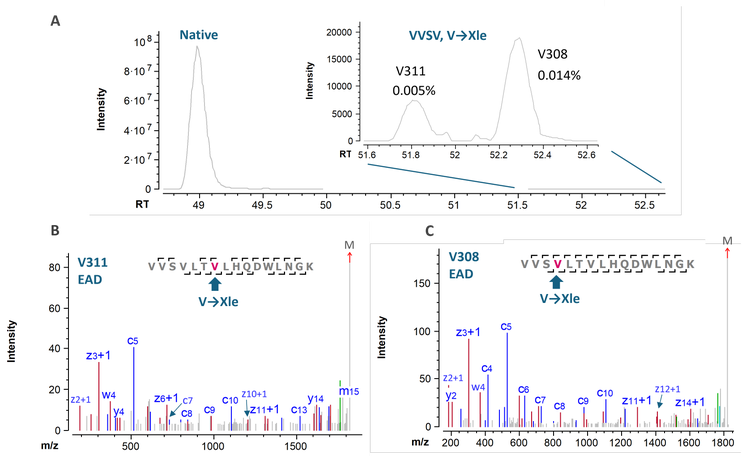
The presence of SVs in biotherapeutics has attracted attention from the biopharmaceutical industry and regulatory agencies.9 SVs are challenging to identify and characterize due to their low levels in biotherapeutics. This challenge can be addressed by the MS sensitivity gain provided by the ZenoTOF 8600 system. Figure 5 shows the XICs and EAD spectra of 2 low-abundant SVs of the VVSV peptide containing the V→Xle (Xle = Ile or Leu) substitution in 2 different positions. Despite the extremely low level of these 2 isomeric SVs (<0.02%), EAD led to extensive sequence fragmentation for their confident identification. The high-quality EAD data also allowed for the differentiation of these 2 positional isomers, where the V→Xle substitution occurs at V308 and V311, respectively.
Disulfide bond mapping
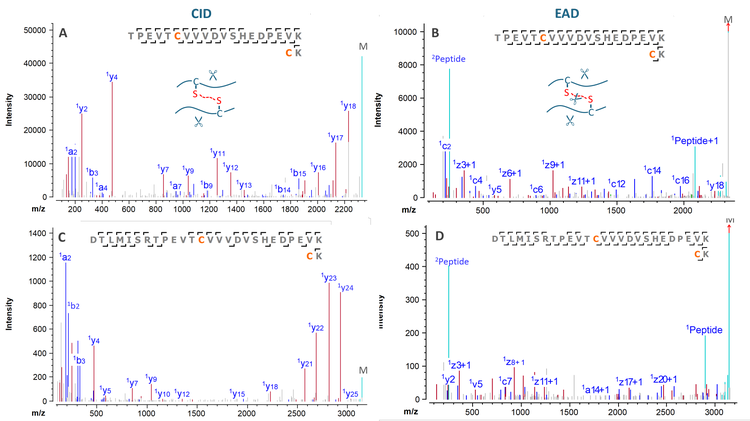
EAD provides complementary and superior fragmentation to CID for disulfide bond mapping. EAD can simultaneously cleave the peptide backbone and the disulfide bond, leading to more extensive fragmentation and increased confidence in sequence identification.8Figure 6 shows the CID and EAD spectra of 2 disulfide-linked peptides from the non-reduced trypsin digest of NISTmAb. While both MS/MS techniques produced high-quality data for confident identification of this peptide, EAD generated more sequence fragments for higher peptide coverage by cleaving the disulfide bond (Figures 6B and 6D). The cleavage of the disulfide bond also led to the detection of 2 intact peptides
(Peptide1 and Peptide2 in Figures 6B and 6D), providing additional mass confirmation of 2 disulfide-linked peptides. The MS sensitivity gain offered by the ZenoTOF 8600 system enabled the sensitive detection and identification of additional low-abundant disulfide-linked peptides, such as the example shown in Figures 6C and 6D, leading to increased confidence in disulfide bond mapping and in-depth characterization of mispaired disulfide linkages.
In summary, the significant improvement in MS sensitivity offered by the ZenoTOF 8600 system enhances the ability of the CID or EAD-based peptide mapping workflow for the detection, identification and characterization of low-abundant PQAs in biotherapeutics. This analytical breakthrough expands the utility of EAD for a wide range of biopharma applications, from routine peptide mapping to advanced PTM analysis, isomer differentiation, disulfide bond mapping and SV analysis.
Conclusion
- The ZenoTOF 8600 system significantly increases MS sensitivity by up to 10X compared to the ZenoTOF 7600 system for enhanced peptide mapping using CID or EAD DDA, even with low sample loads.
- The MS sensitivity improvement compared to the ZenoTOF 7600 system enables the detection and identification of lowabundant PQAs, such as W oxidation species, various PTMs and SVs.
- EAD offers unique capabilities for accurate localization of labile PTMs and unambiguous differentiation of amino acid isomers. The enhanced sensitivity enables precise localization of labile PTM even for trace glycans.
- CID and EAD provide complementary information for confident disulfide bond mapping.
- The enhanced peptide mapping workflow can be employed for routine to advanced characterization of biotherapeutics to ensure drug quality, safety and efficacy.
References
- Anna Robotham and John Kelly. (2020) LC-MS characterization of antibody-based therapeutics: recent highlights and future prospects. Approaches to the Purification, Analysis and Characterization of AntibodyBased Therapeutics. Chapter 1: 1-33.
- A single-injection workflow for enhanced peptide mapping using collision-induced dissociation (CID) and electronactivated dissociation (EAD). SCIEX technical note, MKT28039-A.
- A single-injection workflow for enhanced peptide mapping using collision-induced dissociation (CID) and electronactivated dissociation (EAD). SCIEX technical note, MKT28039-A.
- Comprehensive characterization of O-linked glycosylation in etanercept by electron activated dissociation (EAD).
SCIEX technical note, RUO-MKT-02-14921-A. - Differentiation of leucine and isoleucine for enhanced sequence variant analysis using electron activated dissociation. SCIEX technical note, MKT-30799-A.
- Comprehensive differentiation isomers from forced degradation by electron activation dissociation (EAD). SCIEX technical note, RUO-MKT-02-14730-A.
- Site-specific differentiation of hydroxyproline isomers using electron activated dissociation (EAD). SCIEX technical note, MKT-30610-A.
- Comprehensive characterization of a cysteine-linked antibody-drug conjugate using electron-activated dissociation (EAD). SCIEX technical note, MKT-34414-A.
- Aiming Zhang et al. (2020) A general evidence-based sequence variant control limit for recombinant therapeutic protein development. MABS. 12(1): e1791399


