Abstract
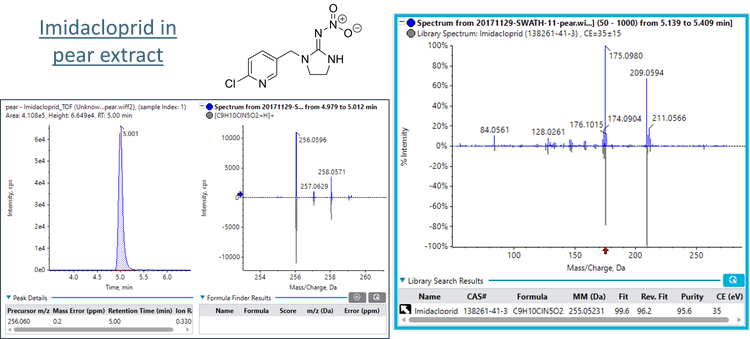
This technical note describes a suspect screening data processing workflow in the SCIEX OS software for the non-targeted analysis of pesticides in fruits, vegetables and herbs. Data was collected using the SCIEX X500R QTOF system and variable-window SWATH data independent acquisition (DIA). The suspect screening list was comprised of 277 pesticides. The SWATH DIA method acquired high-quality MS/MS fragmentation spectra for all precursor compounds. These spectra were compared to the SCIEX Pesticides High Resolution MS/MS Spectral Library to confirm the analyte detection. For example, spectral matching was used to confirm the detection of imidacloprid in pear (Figure 1). The criteria of precursor and fragment mass error, retention time error and isotope ratio were used to provide further confidence in the pesticide detection. Finally, the qualitative flagging rules were used to rapidly and efficiently filter the large data set for positive detections.
Key benefits of using SCIEX OS software for suspect screening of pesticides in food
- Efficient screening for large analyte panels. Combination of SWATH DIA and suspect screening workflow quickly screened food samples for 277 pesticides with high confidence
- Improved compound detection confidence using MS/MS spectral matching. Library searching feature in SCIEX OS software allowed for MS/MS matching to the SCIEX pesticides database to confirm pesticide detection in real-world food samples
- Rapid filtering of Results Table to identify positive hits. Qualitative flagging rules sped up data review by filtering based on criteria of mass error, retention time, isotope pattern and MS/MS library match
Introduction
Non-target acquisition (NTA) methods using high-resolution mass spectrometry (HRMS) instruments, such as the X500 QTOF system, are commonly used for screening large analyte panels in food. Using SWATH data independent acquisition (DIA), high-quality MS/MS fragmentation spectra are collected for all precursor compounds. The MS/MS spectra can be compared against a library database of known chemicals, providing an additional line of evidence to improve compound detection confidence. The positive MS/MS library match is used in addition to criteria such as retention time, isotope ratio and, precursor and fragment ion mass error. However, the processing and review of HRMS data can be time-consuming due to the large amount of data generated. Therefore, it is important for HRMS data processing to be fast, efficient and powerful.
This technical note highlights the data processing workflow for the non-target analysis of pesticides in fruits, vegetables and herbs using a suspect screening list of 277 pesticides. Using the SCIEX OS software, MS/MS library matching was performed with efficient data filtering to quickly identify potential pesticide detections. The combination of SWATH DIA acquisition with suspect screening and SCIEX OS data processing is a rapid and efficient method for identifying pesticide residues in food.
Methods
Sample preparation: Extracts were prepared from a variety of purchased produce and herb samples.
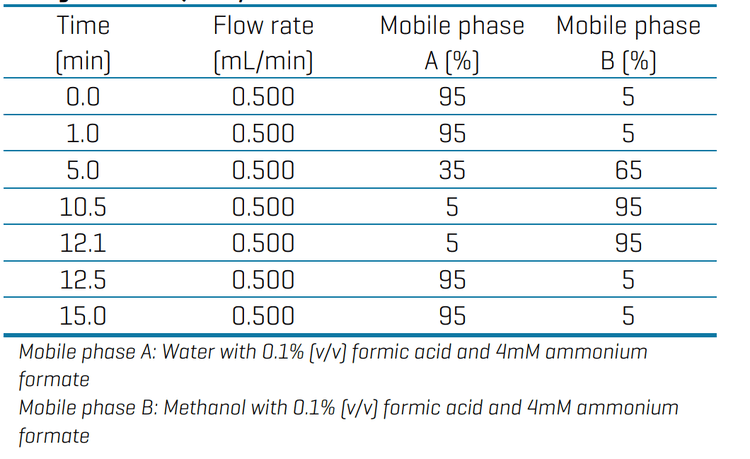
Chromatography: Chromatographic separation was achieved using a SCIEX ExionLC AD system with an Agilent Zorbax Eclipse Plus C18 column (1.8 µm, 4.6 x 50 mm). The mobile phases were water (A) and methanol (B), both modified with 0.1% (v/v) formic acid and 4mM ammonium formate. The runtime was 15 min using the gradient conditions described in Table 1. The flow rate was 0.5 mL/min, the injection volume was 2 µL and the column oven was 400o C.
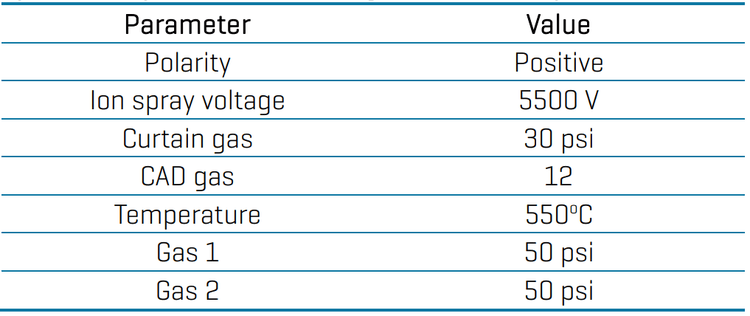
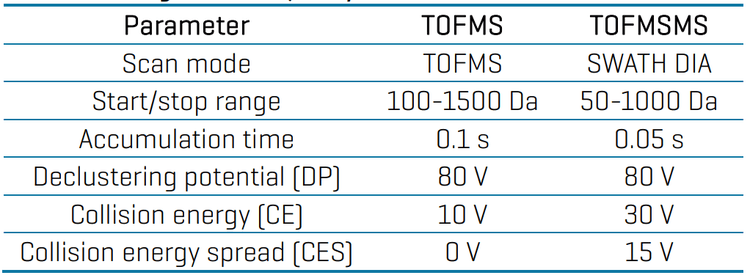
Mass spectrometry: Samples were analyzed using the SCIEX X500R QTOF system in positive electrospray ionization mode. Data was acquired using SWATH DIA with 16 variable-width Q1 windows and an accumulation time of 50 ms. Table 2 presents the optimized source and gas conditions. The TOFMS and TOFMSMS parameters are described in Table 3. In general, the parameters were selected to give optimal sensitivity for most of the targeted pesticides.
Data processing: Samples were processed using the SCIEX OS software (version 3.3). The suspect screening list consisted of 277 pesticides. The components list included the precursor ion and one fragment ion for each suspect compound. The extracted ion chromatogram (XIC) width was 0.02 Da. The SCIEX Pesticides High Resolution MS/MS Spectral Library was used for library searching.
Suspect screening data processing workflow
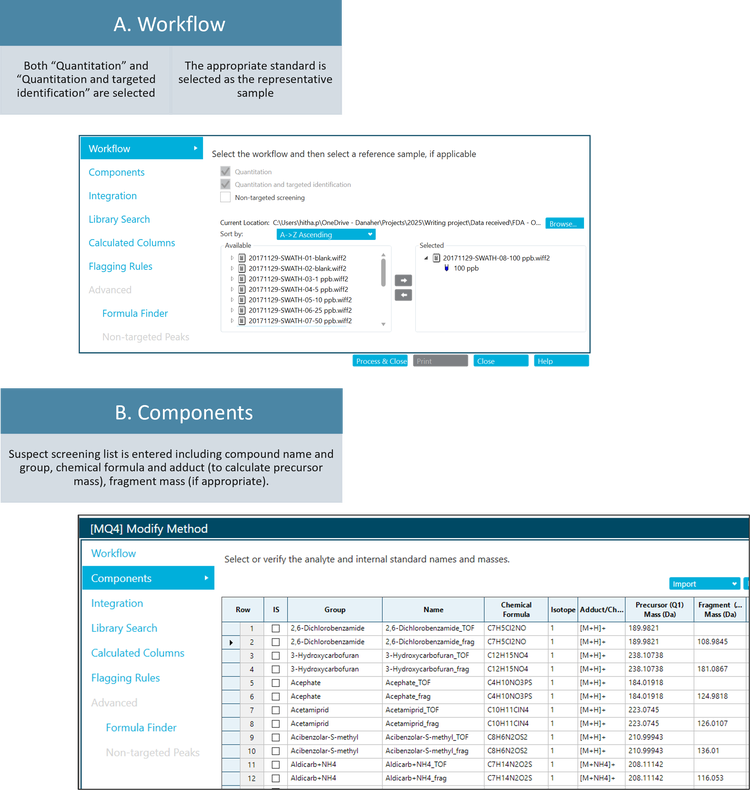
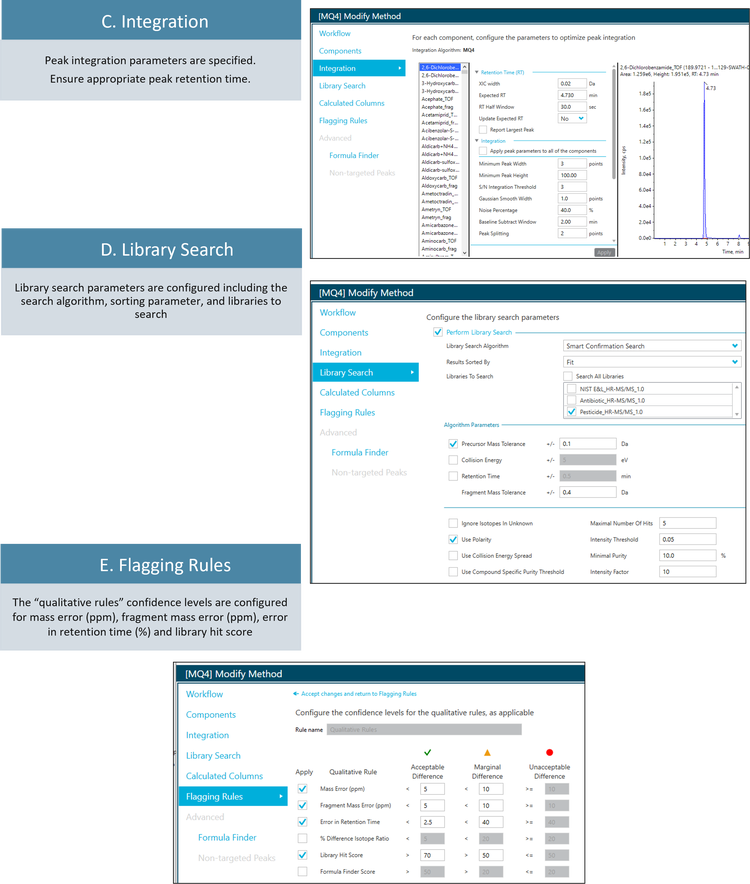
Pesticide screening in real-world food samples
A series of fruits, vegetables and herbs were purchased, extracted and analyzed using SWATH DIA on the X500R QTOF system. In addition to the food extracts, a series of solvent-based calibration standards were run to confirm the analyte retention time and quantify any pesticide detected. In the processing method, the precursor ion and the dominant fragment ion were extracted for each of the 277 pesticides in the suspect screening list (Figure 2). The extraction window was set to 0.02 Da to minimize matrix background in the XIC.
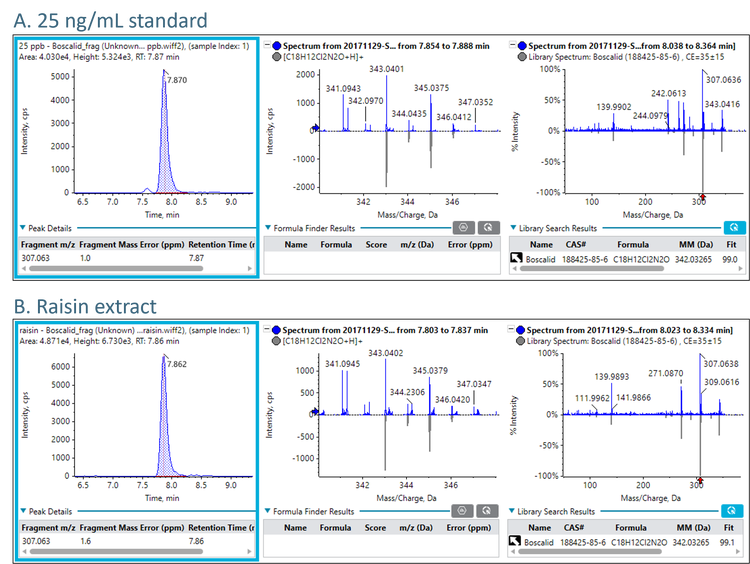
Figure 4 demonstrates the positive detection of boscalid in the raisin sample. For illustrative purposes, Figure 4 only shows the fragment ion XIC, however, the precursor ion was also detected with good mass accuracy (<5 ppm mass error). The top panel (A) shows the 25 ng/mL standard and bottom panel (B) shows the raisin sample extract. Good fragment mass accuracy (<5 ppm mass error) and isotope ratio pattern (middle panels) were observed. The experimental MS/MS spectrum was compared to the database in the SCIEX Pesticides High Resolution MS/MS Spectral Library. The raisin sample correctly matched to boscalid with a very good “Fit” score of 99.1.
In addition, Figure 1 shows the positive detection of imidacloprid in the pear sample extract. In this example, the precursor XIC is shown. Similar to the boscalid in raisin example, good precursor mass error (0.2 ppm) and isotope ratio match were observed. The MS/MS library “Fit” score was 99.6, indicating very good agreement with the database spectrum, and the correct pesticide was matched.
Conclusions
This technical note demonstrated:
- An efficient method for screening a large pesticide panel in food using a combination of SWATH DIA and suspect screening workflow
- Use of the SCIEX Pesticides High Resolution MS/MS Spectral Library for MS/MS matching to improve pesticide detection confidence
- Faster data review through the use of the Qualitative Flagging Rules to rapidly filter the Result Table for positive hits