Abstract
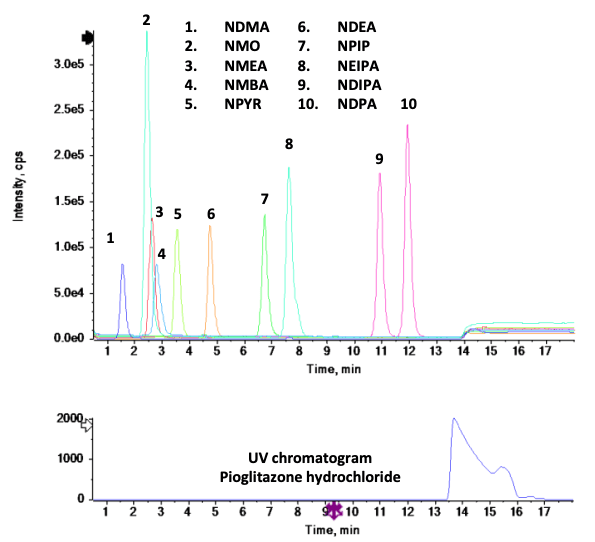
This technical note presents an accurate mass spectrometry method for quantifying 10 mutagenic nitrosamines in Pioglitazone hydrochloride, including NDMA. Excellent chromatographic separation was achieved for all 10 nitrosamines and the Pioglitazone hydrochloride active pharmaceutical ingredient (API) (Figure 1). Statistically significant quantitative performance and linearity were achieved using accurate mass spectrometry at low concentration levels.
Introduction
Pioglitazone is used to treat type 2 diabetes because it increases the effectiveness of insulin produced by the body to help maintain blood sugar levels and alleviate symptoms.5 It is essential to ensure that drug products used to treat disease are free from contamination and safe to use. As a result, medicines such as Pioglitazone have been scrutinized since the nitrosamine crisis began in 2018.4
The recommended limit for total nitrosamines in most drug products is currently 30 ng/g, which is derived from a maximum daily dose of less than 880 mg/day. Pioglitazone hydrochloride has a maximum daily dose of 45 mg and falls well below this threshold where a 30 ng/g limit can be implemented.4
Key features for the analysis of nitrosamines using the X500R QTOF system
- Excellent mass accuracy and selectivity: Achieve low-level quantification of 10 nitrosamines in Pioglitazone hydrochloride API with excellent mass accuracy (<1 ppm) and selectivity
- Rapid acquisition speed: The fast acquisition speed enables the measurement of 10 different nitrosamines simultaneously with precursor ions in full scan TOF MS and MRMHR mode using the X500R QTOF system (Figure 2)
- Statistically significant quantitative performance: Achieve excellent quantitative performance with both precursor ion and MRMHR experiments in terms of linearity, accuracy and precision
- Streamlined data management: Data acquisition and processing are simplified with SCIEX OS software, a 21 CFR Part 11-compliant platform
Methods
Standard preparation: A stock solution containing 10 µg/mL of each nitrosamine was prepared in water from standard solutions. Serial dilutions in water were performed to generate calibration solutions with concentrations of 100, 50, 25, 5, 1, 0.4, 0.2, 0.1, 0.050, 0.025 and 0.010 ng/mL.
Spiked sample preparation: A 100 mg sample of Pioglitazone hydrochloride API was weighed into a suitable vessel. A 5 mL aliquot of a 1 ng/mL nitrosamine mixed standard solution was added and vortexed for 30 seconds. The solution was sonicated for 15 minutes and then centrifuged at 4500 rpm for 5 minutes. The supernatant was removed and filtered through a 0.2 µm PTFE filter and transferred to a HPLC vial for analysis. The resulting solution had a sample concentration of 40 mg/mL with a spike concentration of 1 ng/mL of nitrosamine mix. This was equivalent to the 25 ng/g spike concentration of the sample. 1,2
Chromatography: An ExionLC system with a Phenomenex Kinetex Biphenyl column (2.1 x 100 mm, 2.6 µm, 100 Å) was used for chromatographic separation at a flow rate of 0.4 mL/min. The column was operated at 30°C. Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in methanol. The injection volume was 25 µL. Table 1 summarizes the gradient conditions.
Quantitative performance
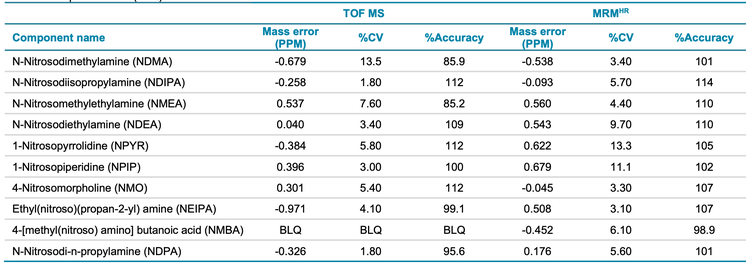
The accuracy of mass measurement is crucial when using an accurate mass spectrometer. This is increasingly important for compounds that have low molecular weights, such as nitrosamines. Consequently, Table 5 shows the high levels of mass accuracy that the X500R QTOF system can achieve with precursor and fragment ions that are used for the quantification of nitrosamines in spiked samples at 1 ng/mL, equivalent to 25 ng/g of Pioglitazone hydrochloride API.
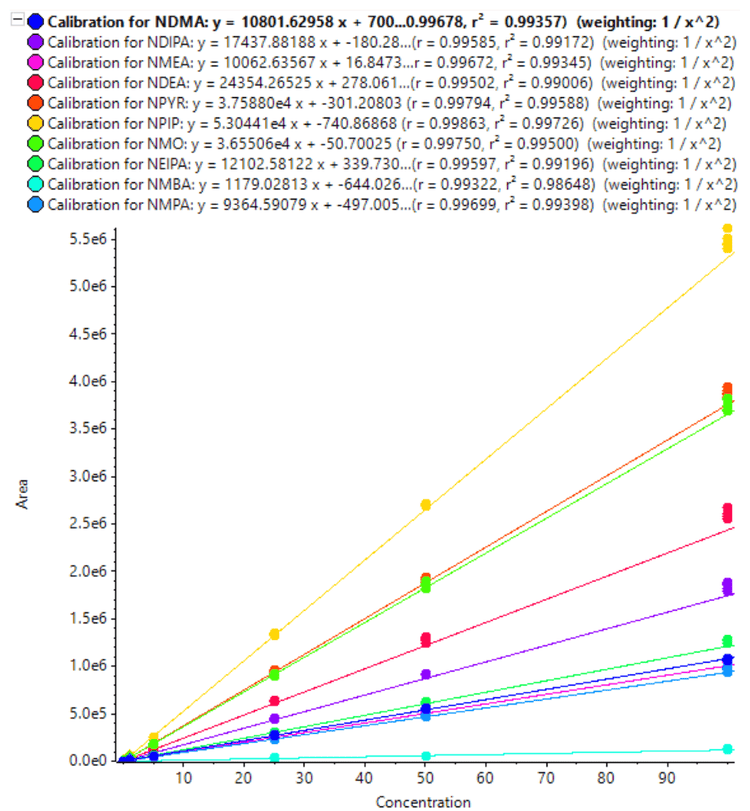
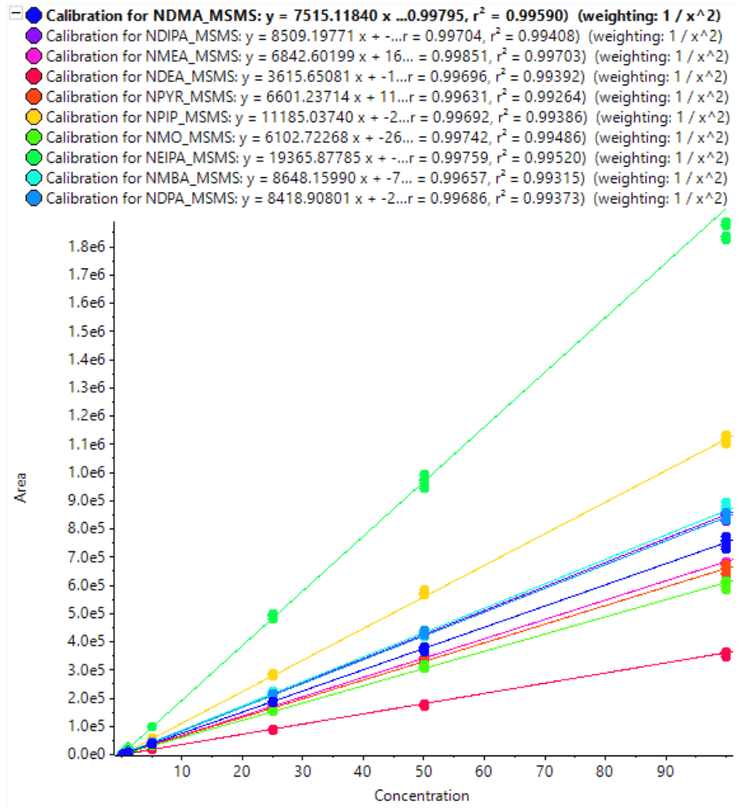
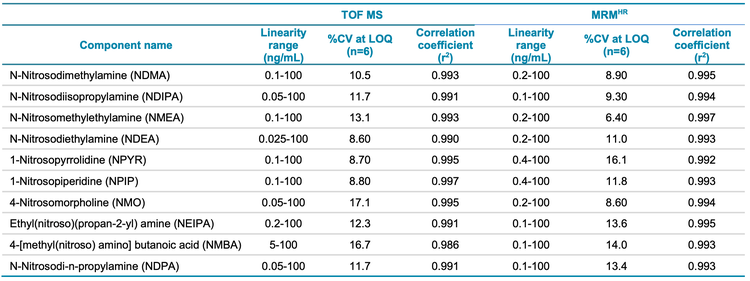
The calibration curves for 10 nitrosamines were plotted across a concentration range of 0.010-100 ng/mL (Figures 3 and 4). A linear dynamic range of 3 orders of magnitude was achieved for most nitrosamines. No carryover was observed within the blank injection following the highest concentration.
A high level of accuracy was achieved across the calibration range, meeting the requirements for nitrosamine impurities in Pioglitazone hydrochloride. For both precursor ion and MRMHR based quantification of nitrosamines, the r2 value was >0.98 (Table 4).
With accurate mass spectrometry, users can choose a workflow that best meets their needs. With TOF MS, method setup is straightforward and requires minimal method development. The MRMHR workflow adds another layer of selectivity with the flexibility to choose the most sensitive and selective fragments for quantification.
Accurate and precise quantification in spiked samples
Accuracy and precision metrics were evaluated in standard solutions and spiked samples. A 1 ng/mL concentration in spiked solution (equivalent to 25 ng/g in sample concentration) was used for the assessment. The acceptable criteria for accuracy and precision at this concentration level were ±30% and <25% of the nominal concentration, respectively.
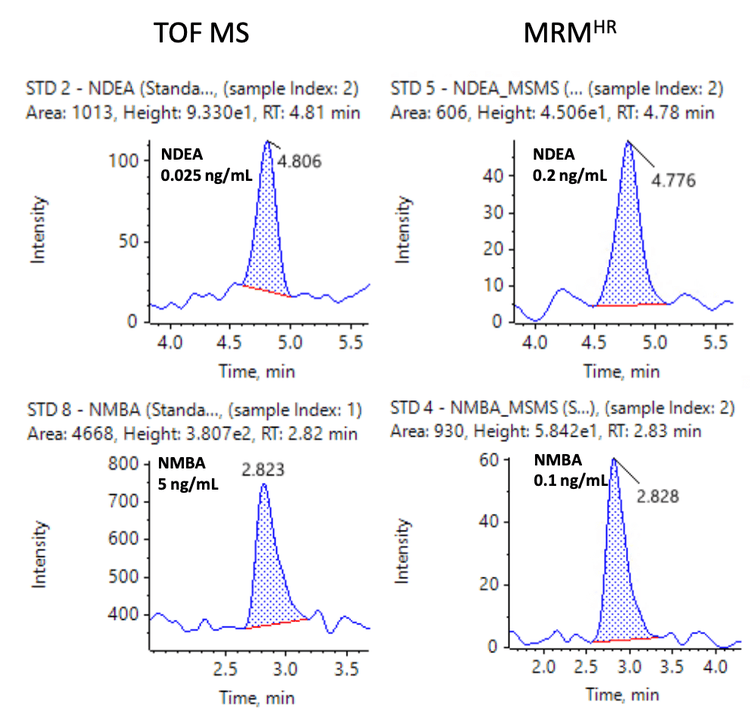
The spiked Pioglitazone hydrochloride API sample met the specified requirements for all nitrosamine impurities (Table 5). Overall, the %CV was <13.5% and <13.3% for precursor ion and MRMHR quantification experiments, respectively. The percent accuracy was within ±15% of the nominal concentration for both quantification workflows. The mass error was <1 ppm in spiked samples, demonstrating high mass accuracy for nitrosamine impurity analysis in the API. The LOQ for NMBA was 5 ng/mL using the precursor ion TOF MS workflow which was found to be BLQ in spiked samples.
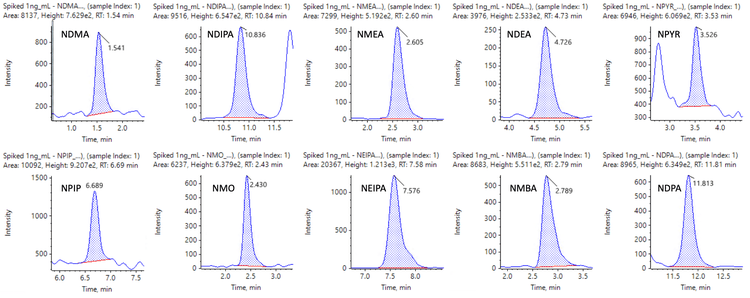
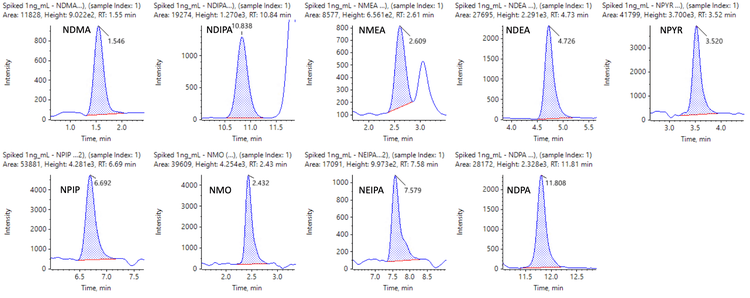
Figures 6 and 7 show representative chromatograms of 10 nitrosamines in 1 ng/mL spiked samples using MRMHR and 9 nitrosamines using TOF MS experiments for quantification.
Compliance-ready SCIEX OS software
To meet the regulation outlined in 21 CFR Part 11, SCIEX OS software is designed as a closed system, and includes the requirement for the records and signatures to be stored electronically. SCIEX OS software can open raw data files from any visible storage location, which enables the flexibility to work within a closed network using designated processing workstations. Figure 8 illustrates 3 types of controls that are required for 21 CFR Part 11 compliance. The workflow presented here is fully compliant with these guidelines, as SCIEX provides 1) technical controls over hardware and software configuration, 2) network security and secure operating systems and policies and 3) procedures and user training (Figure 8).
Conclusion
- Low-level quantification was achieved for 10 nitrosamines in spiked samples using the X500R QTOF system
- Excellent linearity and precision were reached for the analysis of nitrosamines, demonstrating exceptional quantitative performance
- High mass accuracy (<1 ppm) for low molecular weight nitrosamines was accomplished, minimizing false positive results
- Utilizing the fast-scanning speed of the X500R QTOF system, simultaneous monitoring was performed on 10 nitrosamines in precursor ion in full scan TOF MS mode and MRMHR experiments
- The method demonstrated the quantification of nitrosamine impurities below the current recommended limit (30 ng/g) in the Pioglitazone hydrochloride drug product
- A single platform for streamlined data acquisition, processing and management with SCIEX OS software was presented
References
- Analysis of nitrosamine impurities in a metformin drug substance and drug product, SCIEX technical note, RUOMKT-02-12679-A.
- Analysis of genotoxic nitrosamines in losartan using the SCIEX 7500 system, SCIEX technical note, RUO-MKT-02- 13005-B.
- Update on nitrosamine impurities: EMA continues to work to prevent impurities in medicines, 26 April 2019, European Medicines Agency, EMA/241020/2019
- Control of Nitrosamine Impurities in Human Drugs, February 2021, US food and drug administration Guidance for Industry, Revision 1
- National Health Service, Medicines A to Z, 14 March 2022 – Pioglitazone
- Diabetes fact sheet, 16 September 2022, World health organization




