Abstract
MRM3 analysis enables higher specificity peptide detection and therefore better quantification of peptide from highly complex samples. Improved detection of PSA in serum on the QTRAP 5500 System is demonstrated.
Introduction
Over the last ten years, mass-spectrometry based proteomics has been applied to the discovery of putative protein biomarkers of disease from comparably small numbers of samples. However, there has been limited verification / validation of these putative biomarkers and therefore minimal translation of these proteins into assays of clinical utility. Immuno-based assays have been traditionally used for detection and quantification of proteins in clinical research. Developing these tests, however, is time-consuming. Analytical techniques are needed that will enable researchers to develop protein assays rapidly and produce assays with high specificity and sensitivity. This is driving a rapidly growing interest in using LC/MS-based strategies to quantify proteins.
The use of Multiple Reaction Monitoring (MRM) combined with stable isotope labeled proteins / peptides for the quantification of proteins has been actively explored over the last few years and shows great promise for clinical research. Some key requirements of protein assays for clinical research are high specificity, high robustness, very high throughput and the ability to quantify low abundance proteins in the ng/mL to pg/mL concentration range in e.g. human plasma. Also, the sample preparation must be simple and robust.
Another limitation hindering the widespread use of MRM for biomarker verification has been the widespread use of nanoflow chromatography. While it greatly increases the overall sensitivity of LC-MS/MS experiments and requires significantly reduced amounts of sample, nanoflow chromatography does currently not offer the sample throughput, reproducibility and robustness required for implementation in clinical research laboratories.
Here, a novel approach is presented that combines the use of analytical chromatography with a highly selective mass spectrometry technique called MRM3 that is able to significantly reduce sample complexity, available on SCIEX QTRAP Systems. The approach enables robust detection of protein biomarkers from human serum at concentrations down to the low ng/mL level.
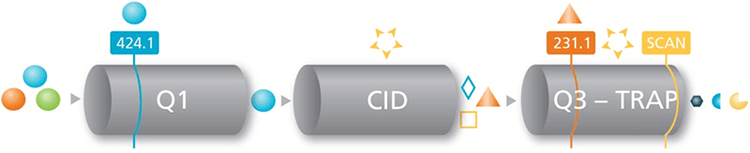
Key features of Using MRM3 for quantifying of protein biomarkers
- Technology advancements in the QTRAP 5500 System3 enables faster, more sensitive MRM3
- MRM3 analysis enables higher specificity peptide detection and therefore better quantification of peptide from highly complex samples
- Higher specificity can also reduce the fractionation required for low level detection and increase sample throughput
Methods
Sample preparation: Human serum samples were denatured with 6M urea, reduced with 30 mM dithiothreitol and alkylated with 50 mM iodoacetamide. Proteins were digested with trypsin overnight at 37 °C (1:30 w/w enzyme to substrate ratio). Samples were desalted using reversed phase cartridges (Oasis HLB 3 cm3, Waters) and fractionated using an MCX cartridge with elution at pH 5.5 using a methanol/acetate buffer mixture (Waters).2
Liquid chromatography: LC-MS/MS analysis was performed using a 2.1 x 100 mm Symmetry C18 reversed phase column (3.5 µm, Waters). A 30 min linear gradient (5-40% acetonitrile in 0.1 % formic acid, 300 µL/min) was used to elute the peptides into the MS system. Total injection to injection cycle time was 45 mins.2
Mass spectrometry: The eluent was analyzed using the Turbo V™ Ion Source on the QTRAP 5500 System in both MRM and MRM3 modes of acquisition3. MRM analysis was performed using Unit resolution both Q1 and Q3 quadrupoles. MRM3 analysis was performed using the MS/MS/MS scan function.2 The precursor ion was isolated in Q1 using unit resolution, first-generation product ions were generated in the Q2 collision cell using an optimized collision energy and trapped in the Q3 linear ion trap for 200 ms. A suitable first-generation product ion was selected and further excited by resonance excitation for 25 ms to produce second-generation fragments. These were finally scanned out of the ion trap at 10 000 Da/sec resulting in a total cycle time of 350 ms per peptide.2 Q0 Trapping was used to further increase sensitivity.
Data processing: Data was processed using MultiQuant™ Software. MRM peak areas were integrated, either individually or summed together. MRM3 peak areas were determined by summing the integration of up to four granddaughter ions.
Previous work showed low level detection of PSA
Recently, the detection of PSA in human serum was demonstrated using LC-MRM analysis on a 4000 QTRAP System using high flow chromatography.1 This method used HSA depletion followed by mixed cation exchange fractionation and a HPLC separation to enable low level detection (Figure 3). As depletion of high abundant proteins is the most difficult step to automate, exploring additional strategies to remove this step are key workflow improvements. Here, the use of MRM3 analysis(Figure 2) has been explored for the detection of PSA and other low abundance proteins in human serum.
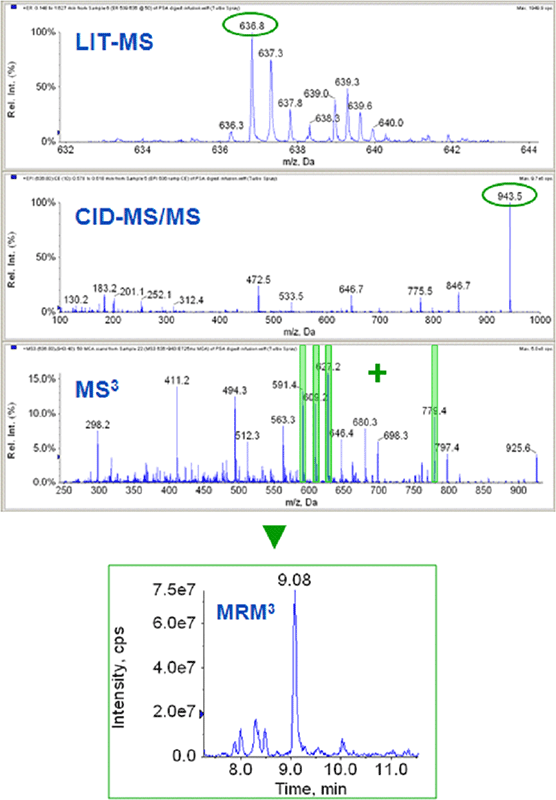
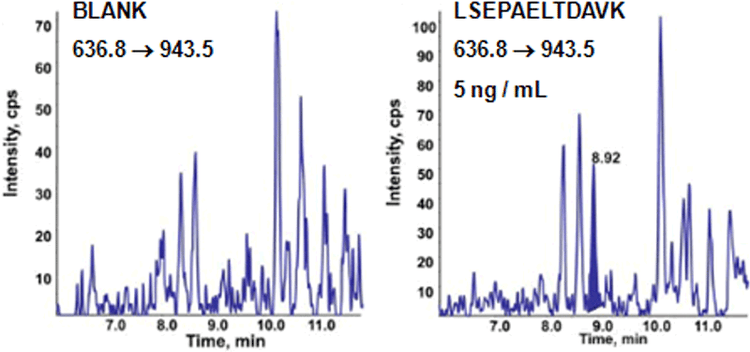
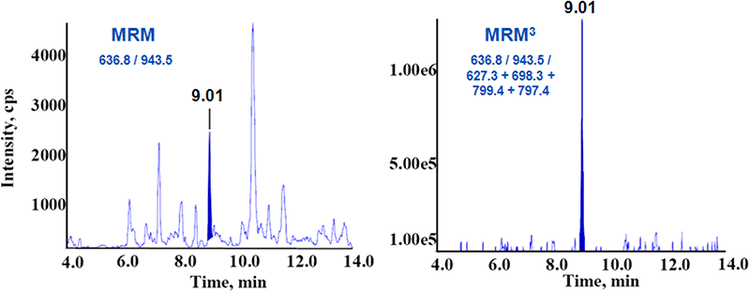
MRM3 for PSA quantification
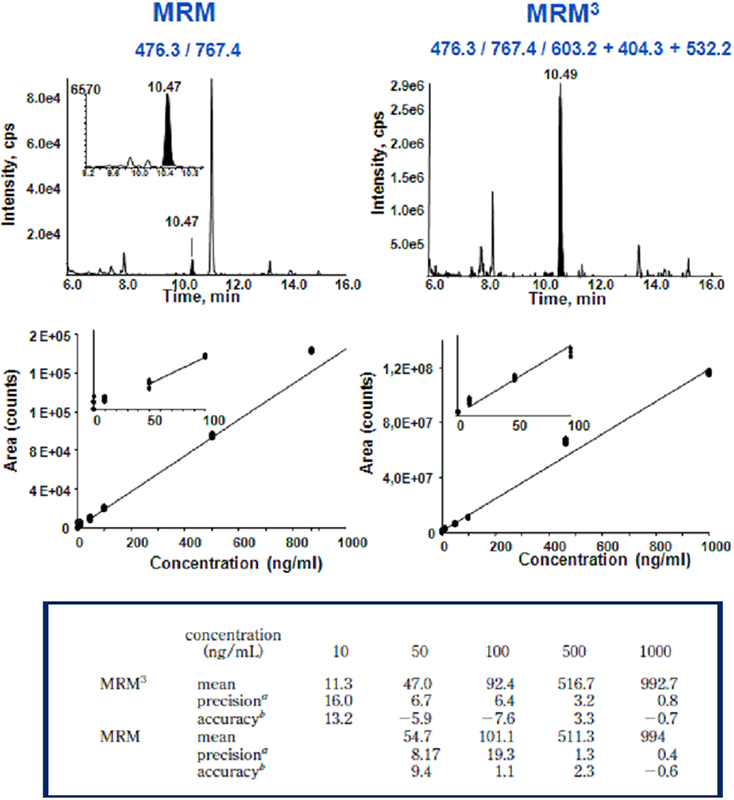
To assess first the intrinsic gain in specificity of detection of the MRM3 method compared to the conventional MRM operating mode, a trypsin digest of a human female serum was spiked with trypsin hydrolyzed bacterial protein models TP171, TP574, TP435, core NS4, and human prostate specific antigen (PSA) over three decades of concentration, ranging from 0 to 1000 ng/mL. On average, moving from MRM to MRM3 resulted in a 3- to 5-fold improvement of the limits of detection and quantification for the five model proteins2. Removal of background interferences / noise is illustrated for PSA in Figure 4. The standard concentration curve data from CoreNS4 protein illustrates the typical improvement (Figure 5). The reduction in background in the LC chromatograms of the MRM and MRM3 data can be observed in the top panes (Figure 5). The middle panes show the standard concentration curves using both acquisition techniques. The table in the lower pane shows the precision and accuracy for both concentration curves, highlighting the improvements in LLOQ achieved by the more selective MRM3 experiment over the standard MRM.
In order to reach the desired limit of quantitation of 4 ng/mL, we applied a previously optimized proteotypic peptide enrichment based on mixed-cation exchange solid phase extraction on a cartridge format just after trypsin hydrolysis of the crude sera2. This extraction phase could also be easily implemented online using an additional chromatographic module. This extraction step combined with MRM3 analysis enabled the removal of the immune-depletion step from the sample preparation strategy.
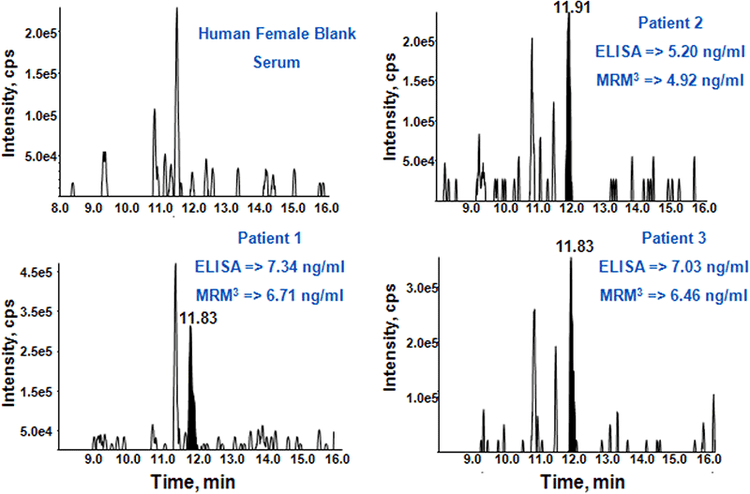
Strong correlation with ELISA assays
The MRM3 quantitation assay was performed on blank female serum and three male samples with known prostate cancer. The measured values of PSA obtained correlated well with the values obtained from the established ELISA assay (Figure 6).2 This clearly demonstrates the potential of the MRM3 approach for the detection and accurate quantification of low-level protein biomarkers in body fluids.
Conclusions
- Quantification of proteins in human serum at low ng/mL concentration levels has been demonstrated using high flow chromatography and LC-MRM3 analysis.
- Critical detection levels of 4-10 ng/mL of circulating PSA were accurately determined in human serum using a robust two step sample preparation protocol combined with LC-MRM3 analysis
- Improvements in speed and sensitivity to the QTRAP 5500 System make MRM3 analysis a robust quantitative strategy for peptides in complex matrices when significant background / interferences are present.
References
- Fortin T et al. (2009) Clinical quantitation of prostate-specific antigen biomarker in the low nanogram/milliliter range by conventional bore liquid chromatography-tandem mass spectrometry (multiple reaction monitoring) coupling and correlation with ELISA tests. Mol. Cell. Prot. 8, 1006.
- Fortin T et al. (2009) Multiple reaction monitoring cubed for protein quantitation at the low nanogram/milliliter level in nondepleted human serum. Anal. Chem. 15, 9343.
- MRM3 quantitation for highest specificity in complex matrices. SCIEX technical note RUO-MKT-02-2739-A.
Related content
- High selectivity quantification of protein isoforms using MRM3 Workflow. SCIEX technical note RUO-MKT-02-4845-A.
- Powerful Scan Modes of QTRAP System Technology. SCIEX technical note RUO-MKT-02-8611-A.
