Automated peptide fragment ion selection and summation for streamlined HRMS quantitative workflows
Abstract
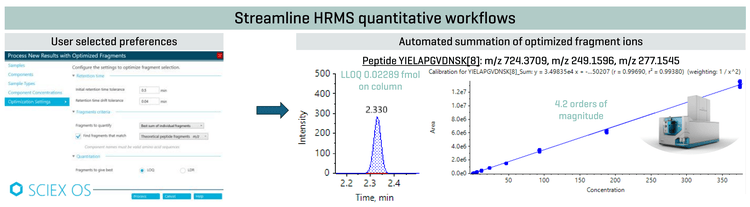
This technical note demonstrates an automated fragment selection workflow to streamline HRMS-based peptide quantitation using SCIEX OS software on the ZenoTOF 8600 system. The workflow enables the automatic selection and summation of optimal peptide fragments based on user-defined criteria, either the lower limit of quantitation (LLOQ) or linear dynamic range (LDR) (Figure 1). Summed fragment ions improved sensitivity for better quantitative performance for peptide workflows.
HRMS has become an essential platform for peptide quantitation, given the increased selectivity resulting in better sensitivity as well as flexibility to generate and monitor a broad range of fragment ions. The accessibility of TOF MS/MS data offers a distinct advantage: users can make post-acquisition decisions to select the most suitable fragments for quantitation, optimizing either sensitivity or dynamic range.
Quantitation can be performed by either using individual fragment ions or summing multiple highly abundant fragment ions. When multiple high abundant fragment ions are generated from the target peptide, the sum of the extracted ion chromatograms (XICs) can further enhance the assay sensitivity.
Often, selecting and summing can be time-consuming and require extensive manual effort. Here, an automated fragment selection and summation approach for peptide quantitation is demonstrated, highlighting its use in deriving optimal fragment ions for best quantitative performance.
Key benefits for HRMS-based peptide quantitation using automated fragment selection with SCIEX OS software on the ZenoTOF 8600 system
- Streamlined HRMS data processing: Automate summation of peptide fragment ions based on user-selected preferences using the automated fragment selection feature on SCIEX OS software
- Enhanced sensitivity with summed fragment ions: Improve LLOQs for peptide quantitation by summing multiple highly abundant fragment ions with the availability of full scan TOF MS/MS data
- Effortlessly meet critical quantitative performance criteria: Achieve accurate quantitative performance with %CV <13 across a wide range of concentrations
- Streamlined data management: SCIEX OS software, a 21 CFR Part 11-compliant platform, simplifies data acquisition and processing
Introduction
Peptide quantitation plays a crucial role in biopharmaceutical research in driving insights into biomarker discovery, pharmacokinetics and drug safety and efficacy. HRMS has emerged as a preferred platform for peptide quantitation, owing to its increased selectivity and sensitivity, as well as flexibility in fragment ion generation.
For quantitative applications using HRMS, maximizing performance often requires a thorough selection and summation of fragment ions, a process that can be highly time-consuming.
Here, an automated fragment selection workflow is demonstrated, highlighting a simplified approach that accelerates peptide quantitation by automatically selecting optimal fragment ions based on user-defined criteria. By fully leveraging the rich information available in TOF MS/MS data using the ZenoTOF 8600 system, this streamlined workflow enhances sensitivity, offering a more efficient path to high-quality quantitative results.1
Methods
Sample preparation: Protein precipitation was performed using a 1:3 (v/v) plasma/acetonitrile ratio. Samples were vortexed and centrifuged at room temperature. The supernatant was further diluted at a 1:4 (v/v) supernatant/water (water containing 2% acetonitrile, 2% acetic acid and 0.1% formic acid). A mixture of 25 peptides was spiked into diluted rat plasma extract and serially diluted to create a standard curve.
Chromatography: Sample separation was performed using Phenomenex Kinetex XB C18 column (2.1 x 50 mm, 2.6 μm) on an ExionLC AD system (SCIEX) with a flow rate of 0.8 mL/min. For separation, a 5-minute method with 0.1% formic acid in water as mobile phase A and 0.1% formic acid in acetonitrile as mobile phase B (Table 1) was used. The column was kept at 40°C. An injection volume of 5 μL was used for analysis. A mixture of 1:1:1 (v/v/v), acetonitrile/methanol/water was used as a needle wash solvent.
Mass spectrometry: Samples were analyzed using a ZenoTOF 8600 system (SCIEX) operating in positive ion mode. The data were acquired using a Zeno MRMHR experiment. The optimized MS parameters are listed in Table 2.
Data processing: Analysis was performed using SCIEX OS software, version 4.0 (SCIEX). Peaks were integrated using the MQ4 algorithm, and a weighting of 1/x2 was used to quantify all peptides in the peptide mix. An XIC peak width of 0.02 Da was applied for quantitation.
Summation of multiple fragment ions enhances sensitivity
The accessibility of full scan TOF MS/MS data can be advantageous as post-acquisition data decisions can be made on which measured fragments can be utilized for MRMHR. For MRMHR, quantitation can be performed using single fragment ion or by summing multiple dominant fragment ions. When multiple high-abundant fragment ions are generated from the target peptide, the sum of XICs can further enhance the assay sensitivity.
Often, the process of selecting and summing fragment ions can require a lot of manual work upfront. Here, the use of the automated selection of fragment ions feature on SCIEX OS software was demonstrated. Figure 1 shows the user-based selection criteria in the Analytics module.
Under the optimization settings, the user can choose preferences for retention time, fragments and quantitation settings. Under retention time, the initial retention time tolerance as well as the retention time drift tolerance can be configured. The fragments criteria allow users to select whether to sum fragment ions or use individual fragments, as well as align fragment ions with either theoretical values or from a library. For the results shown in this technical note, the best sum of individual fragments was selected by the algorithm, where matching was performed with theoretical peptide fragments. Additionally, the user can select a choice between fragments that give the best LLOQ or fragments that give the best LDR. In this study, fragments that generate the best LLOQ were chosen.
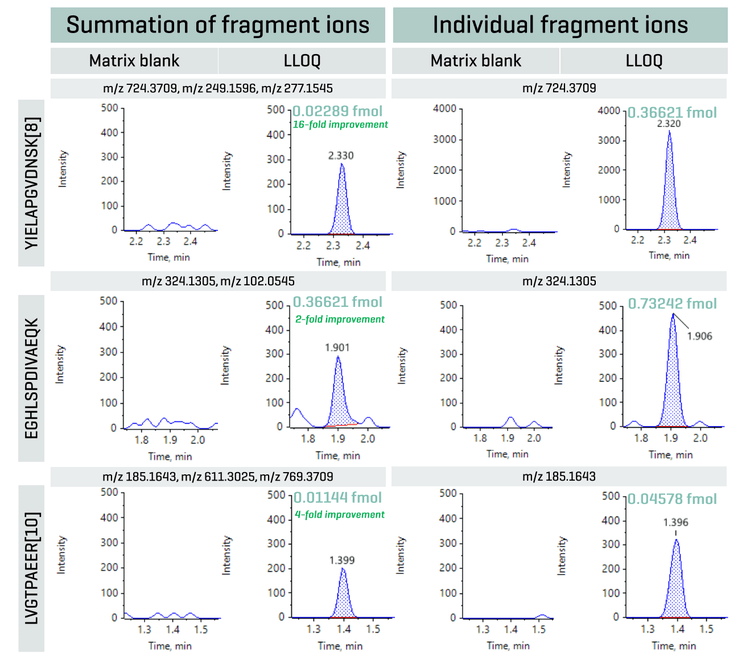
As shown in Figure 2, summing multiple dominant fragment ions can improve the overall LLOQ. No matrix interferences were observed in the blank XICs. Peptide YIELAPGVDNSK[8] showed a 16-fold improvement in LLOQ in comparison to the use of individual fragment ions. Here, a total of 3 fragment ions were summed: m/z 724.3709, m/z 249.1596 and m/z 277.1545. Similarly, summation of m/z 324.1305 and m/z 102.0545 led to a 2-fold improvement in LLOQ for peptide EGHLSPDIVAEQK. For peptide LVGTPAEER[10], fragment ions m/z 185.1643, m/z 611.3025 and m/z 769.3709 were summed, enhancing the final LLOQ by 4-fold.
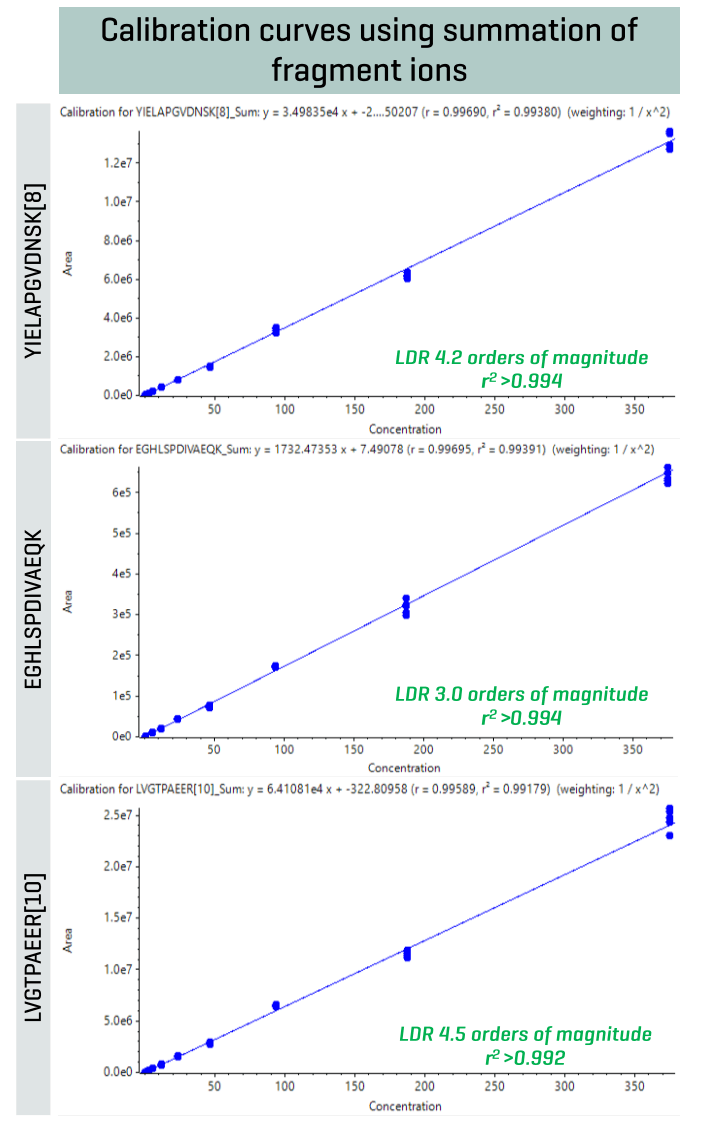
Excellent linearity was observed for the quantitation of peptides using multiple fragment ions (Figure 3). Overall, an LDR of ≥3 orders of magnitude was observed with a coefficient of determination (r2) of ≥0.992 for the 3 example peptides.
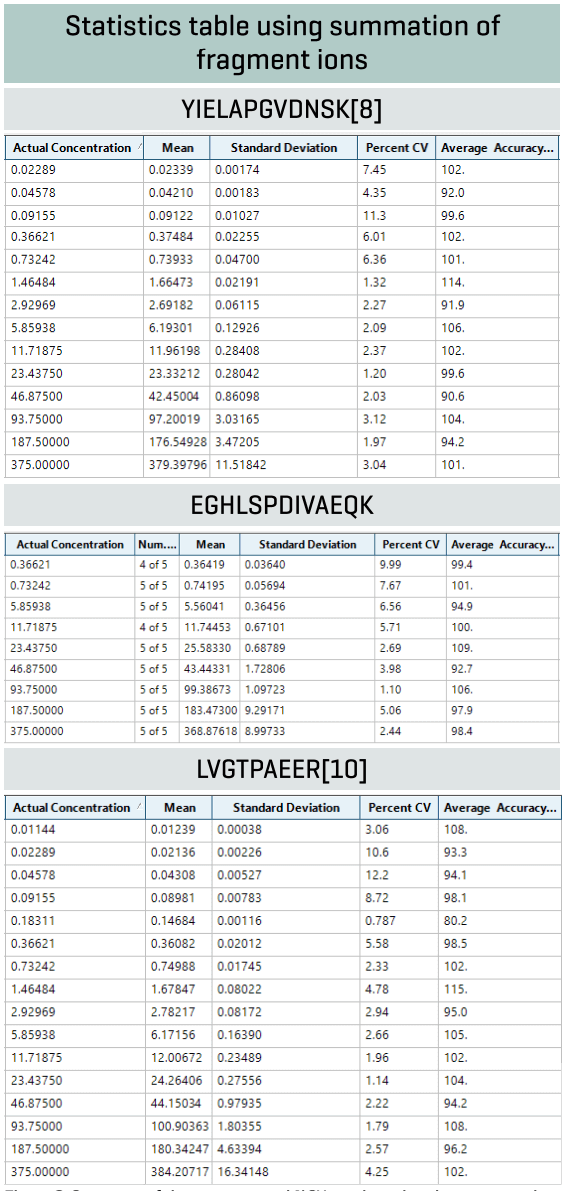
Analytical performance was evaluated based on the requirement that the accuracy of the calculated mean should be between 80% and 120% at the LLOQ and between 85% and 115% at higher concentrations. The %CV of the calculated mean of the concentration should be below 20% at the LLOQ and below 15% at all higher concentrations.2 Accuracy at the LLOQ and at non-zero calibrators was within 85%-115% of the nominal concentration. Overall, precision was <13%, demonstrating high reproducibility (Figure 4).
Overall, improved LLOQs were achieved by summing multiple highly abundant fragment ions, given the availability of TOF MS/MS data. The use of automated fragment selection on SCIEX OS software enabled a streamlined solution for HRMS-based peptide quantitation workflows, leveraging the cumulative gain from the sensitivity provided by the Zeno trap3,4 as well as the summation of highly abundant fragment ions
Conclusion
-
Easily automate summation of peptide fragment ions based on user-selected preferences using the automated fragment selection feature using SCIEX OS software.
-
Improve LLOQs for peptide quantitation by summing multiple highly abundant fragment ions with the availability of TOF MS/MS data.
-
Perform accurate quantitative performance with high reproducibility (%CV <13) across a wide range of concentrations.
-
Retain data management and compliance-readiness (21 CFR Part 11) features using SCIEX OS software to support regulated bioanalysis on the ZenoTOF 8600 system
References
-
Enhanced sensitivity and quantitative performance featuring a novel quadrupole time of flight mass spectrometer. SCIEX technical note, MKT-33800-A.
2. Bioanalytical Method Validation, May 2018.
3. Enabling new levels of quantification. SCIEX technical note, RUO-MKT-02-11886-A.
4. Enhanced sensitivity for peptide quantification in a complex matrix using high-resolution LC-MS/MS. SCIEX technical note, RUO-MKT-02-13324-A.

