Abstract
This technical note highlights the power of a single-injection, EAD-based middle-down workflow for rapid disulfide bond mapping of biotherapeutics with high confidence and high fidelity. The mapping of disulfide bonds on the subunit level minimizes the chance of disulfide scrambling, reduces ambiguities or false positives in determining the disulfide linkages and significantly simplifies data interpretation. EAD led to a characteristic fragmentation pattern of disulfide-linked subunits to rapidly confirm or elucidate the expected or unexpected disulfide linkages with high confidence.
Introduction
Disulfide bonds play a key role in determining the folding, stability and function of biotherapeutics, such as monoclonal antibodies (mAbs). 1 Incorrect formation of disulfide bridges can significantly reduce the efficacy of a biotherapeutic product. 1 Therefore, confirming the disulfide linkages in biotherapeutics is critical to ensure product quality, safety and efficacy. Disulfide bond mapping is traditionally performed using a peptide mapping approach under non-reduced conditions. However, this strategy might lead to ambiguities or false positives in identification due to disulfide scrambling and/or poor MS/MS spectra of low-abundant disulfide-linked peptides. By comparison, the EAD-based middle-down workflow2-5 provides a clear and direct mapping of intra-chain disulfide linkages on the subunit level (Figure 1) without the challenges mentioned above.

Key features of the EAD-based middle-down workflow for disulfide bond mapping
- High confidence and high fidelity: EAD leads to a distinctive fragmentation pattern of disulfide-linked subunits for disulfide bond mapping with minimal false positives
- Minimal disulfide scrambling: Disulfide bond mapping on the subunit level reduces disulfide scrambling and eliminates the need to identify low-abundant disulfide-linked peptides, significantly increasing confidence in mapping results
- Single-injection method: The workflow provides high-quality middle-down data in a single injection
- Streamlined and easy to implement: The workflow requires minimal method optimization and is streamlined from data acquisition to results review
- Powerful software tools: Biologics Explorer software provides optimized workflow templates for improved user experience with data analysis and powerful visualization tools for results review and comparison
Methods
Sample preparation: An aliquot of 10 µg/µL of NISTmAb was diluted in water to a concentration of 1 µg/µL. Then, 50 units/µL of FabRICATOR (IdeS) from Genovis and 50mM dithiothreitol (DTT) were added. The NISTmAb mixture was incubated at 37°C for 15 minutes. For trispecific antibody (tsAb), the sample was first treated with GlySERIAS (Genovis) for 2 days at 37°C, then incubated with 20mM DTT for 5 minutes at 37°C. The reduced tsAb subunits were alkylated for 30 minutes at room temperature using 40mM iodoacetamide. Finally, 5 µL of the NISTmAb or tsAb solution (~5 µg) was injected for LC-MS analysis.
Chromatography: The subunits of NISTmAb and tsAb were separated using an ACQUITY UPLC Protein BEH C4 column (2.1 mm × 50 mm, 1.7 µm, 300 Å, Waters). The LC gradient used for the subunit separation is shown in Table 1. A flow rate of 0.3 mL/min was used for all LC runs. The column was kept at 60°C in the column oven of an ExionLC system (SCIEX). Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile.
Mass spectrometry: MRMHR EAD experiments were performed in SCIEX OS software using the ZenoTOF 7600 system. One charge state (15+ or 17+) per subunit was targeted for EAD fragmentation. The key TOF MS and MRMHR settings used are listed in Tables 2 and 3, respectively.
Data processing: MRMHR data were analyzed using a middle-down workflow template in Biologics Explorer software.
Partial reduction of NISTmAb and tsAb
The single-injection, EAD-based middle-down workflow provides consistently high sequence coverages (70%-85%) and accurate localization of PTMs for the fully reduced mAb subunits. 2-5 This powerful workflow was leveraged here to gain insights into the intra-chain disulfide linkages in NISTmAb and a tsAb. To generate disulfide-linked subunits, NISTmAb and tsAb samples were treated with IdeS and GlySERIAS proteases, respectively, then reduced with DTT for a short period of time (5-15 minutes). Under these conditions, the inter-chain disulfide bonds were reduced while the intra-chain linkages remained intact.
For NISTmAb, IdeS treatment and partial reduction led to the formation of LC, Fd and Fc/2 subunits containing 2 intra-chain disulfide bonds (Figure 2A). The LC and Fd subunits also contained 1 and 3 free Cys residues, respectively, from the reduction of inter-chain disulfide bonds. The 3 disulfide-linked subunits were chromatographically separated in an elution order similar to their fully reduced counterparts. 2,3 Accurate mass measurement of these subunits using Biologics Explorer software confirmed the presence of 2 disulfide bonds (-4 Da) in each subunit (Figure 2C). Similarly, intact mass analysis of the partially reduced tsAb sample revealed the presence of 2 disulfide bonds and 1 carbamidomethyl group on the LC subunit (data not shown). To determine how disulfide bonds are connected in these subunits, a representative charge state (15+ or 17+) of each subunit was targeted for EAD fragmentation, as will be described below.
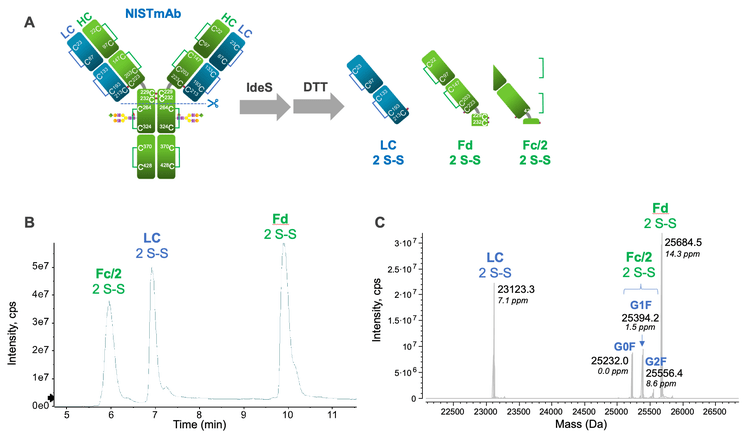
EAD-based middle-down analysis of disulfide-linked subunits
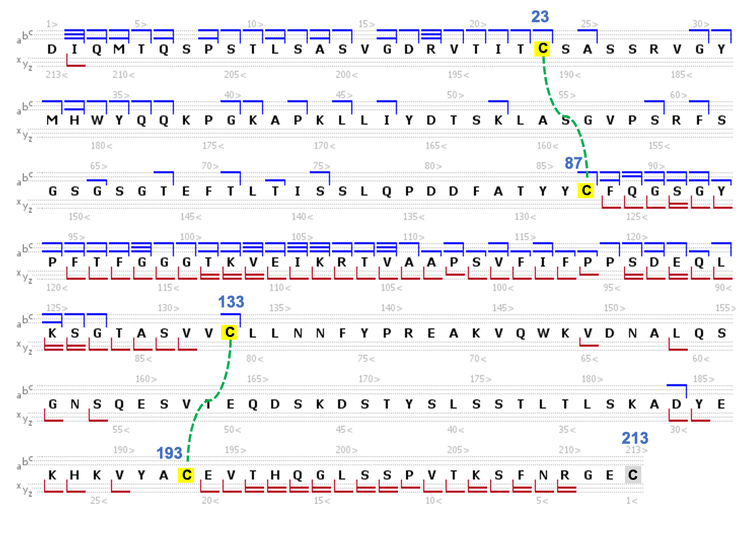
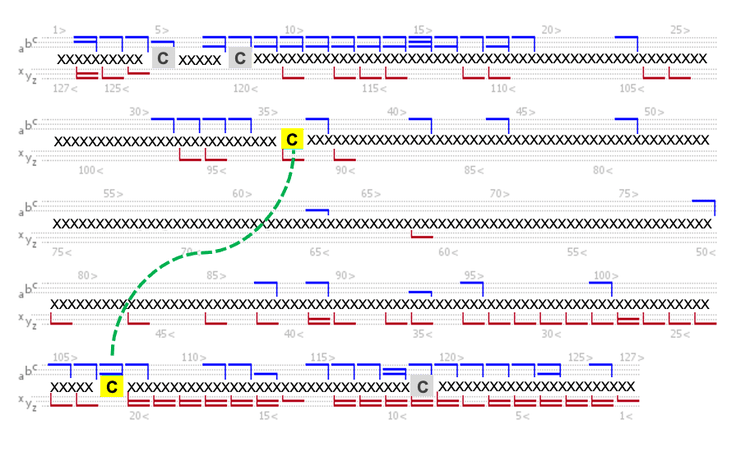
Figure 3 shows an annotated EAD spectrum of the LC subunit containing 2 intra-chain disulfide bonds. The spectrum is displayed in 3 m/z ranges for a better view of rich fragments across different mass ranges. It was described previously that EAD of the fully reduced LC subunit led to a relatively even distribution of c/z fragments from the first or second half of the sequence.2-5 By comparison, the fragments detected in EAD of the disulfide-linked LC subunit were predominantly from the regions not restricted by the disulfide bridges (Figure 3).
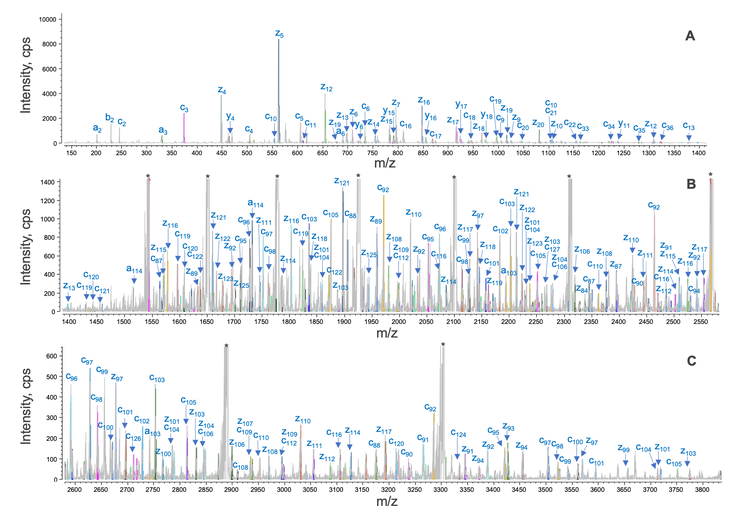
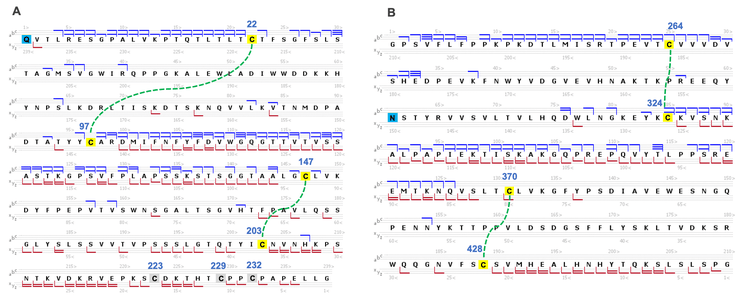
Specifically, a series of large c/z fragments at m/z values >1500 (Figures 3B and 3C) were produced from the backbone cleavages between the Cys87 and Cys133 residues, as depicted in the sequence coverage map shown in Figure 4. The fragments detected at m/z values <1500 (Figure 3A) are mainly from the N- or C-terminal regions beyond the Cys23 and Cys193 residues (Figure 4). A similar fragmentation pattern of the disulfide-linked LC subunit was also observed for the Fd and Fc/2 subunits (Figure 5). These results show that disulfide bridges restricted the fragmentation to the regions not ‘protected’ by the disulfide bridges. The characteristic EAD fragmentation pattern of the disulfide-linked subunits can be leveraged to confirm the expected disulfide linkages or elucidate the mispaired connections.
The powerful capability of the EAD-based middle-down workflow for disulfide bond mapping was employed to characterize a partially reduced tsAb treated with GlySERIAS. Figure 6 shows the EAD spectrum and sequence coverage map of the disulfide-linked LC subunit from the tsAb. Similar to the NISTmAb results described above (Figures 3-5), EAD provided excellent fragmentation of the sequence regions that were not covered by the disulfide bridges, allowing for confident confirmation of the disulfide connections in this LC subunit. In another example, the disulfide linkage in a tsAb subunit (~13.7 kDa) from GlySERIAS cleavage of the HC1 subunit was confirmed based on the detection of the fragments primarily from the N- and C-terminal ends of the sequence (Figure 7). EAD of the disulfide-linked subunits revealed that the disulfide bond was reduced in some fragments detected in the high m/z range, resulting in a 2 Da increase in their masses. Similar results were obtained for the fragmentation of disulfide-linked peptides. 6,7 The mechanism for the formation of these fragments will be discussed in a future technical note.

Conclusion
- A streamlined, EAD-based middle-down workflow led to a characteristic fragmentation pattern of disulfide-linked subunits for rapid disulfide bond mapping with high confidence and high fidelity
- This workflow minimizes the chance of disulfide scrambling and reduces the ambiguities or false positives in determining the intra-chain disulfide bridges
- EAD produced rich fragments across the full m/z range from the sequence regions not restricted by the disulfide bridges
- The EAD-based middle-down workflow combines accurate mass measurement, reproducible EAD fragmentation and automated data analysis and provides an in-depth characterization of fully or partially reduced biotherapeutics
- Biologics Explorer software offers easy-to-use middle-down workflow templates and powerful visualization tools for improved user experience with data analysis
References
- Xiaoyuan Guan et al. (2018) Direct mass spectrometric characterization of disulfide linkages. mAbs. 10(4): 572-582.
- A streamlined single-injection middle-down workflow using electron activated dissociation (EAD) for biotherapeutics characterization. SCIEX technical note, MKT-26997-A.
- Obtaining high sequence coverage and confident posttranslational modification (PTM) analysis of biotherapeutics using an electron activated dissociation (EAD)-based middle-down workflow. SCIEX technical note, MKT-27223- A.
- Comparative analysis of biotherapeutics using an electronactivated dissociation (EAD)-based middle-down workflow. SCIEX technical note, MKT-27427-A.
- Confident sequence analysis of a trispecific antibody using an electron-activated dissociation (EAD)-based middle-down workflow. SCIEX technical note, MKT-27784-A.
- Comprehensive characterization of complex linkage structures in a bispecific monoclonal antibody (mAb) using electron activated dissociation (EAD). SCIEX technical note, RUO-MKT-02-14373-A.
- Comprehensive mapping of disulfide linkages in etanercept using an electron activated dissociation (EAD) based LCMS/MS methodology. SCIEX technical note, RUO-MKT02-15195-A.


