Comprehensive mapping of disulfide linkages in etanercept using an electron activated dissociation (EAD) based LCMS/MS methodology
Featuring the ZenoTOF 7600 system and Biologics Explorer
Zhengwei Chen and Lei Xiong
SCIEX, USA
Abstract
The technical note describes the comprehensive mapping of disulfide linkages in etanercept utilizing the EAD approach on the ZenoTOF 7600 system. The result showcases EAD as a powerful tool providing solid sequence and disulfide linkages confirmation superior to CID, especially for long disulfide peptides with 2 or more disulfide linkages. EAD also preserves the labile O-glycan, enabling site-specific characterization of O-glycosylated disulfide-bonded peptides.
Introduction
Disulfide linkages in biotherapeutic proteins impact their conformation and therefore are considered critical quality attributes (CQAs). LC-MS-based analytical characterization is vital to confirm disulfide linkages and the sequence of the associated peptides. Etanercept contains 26 intra-chain and 3 inter-chain disulfide bonds. An extremely complex and large disulfide peptide with multiple disulfide linkages can be formed across multiple cysteines, even using multi-enzyme digestion. For example, 4 peptides can be linked by 3 disulfide bonds to create a large disulfide cluster. Traditional CID-based fragmentation provides limited sequence information for such peptides. The ability to use a hot electron-capture dissociation (ECD) method, such as EAD,1,2 to analyze disulfide bond dissociation was investigated in previous study. This method allowed confident sequencing of every peptide involved in the disulfide linkage. For data processing, the Biologics Explorer software offers accurate annotation power by providing effective fragment annotations, including the a/x, b/y and c/z ions that are generated from EAD. For the disulfide peptides that contained more than 2 peptides, the MS/MS spectrum of every peptide was well annotated, allowing solid sequencing of the peptide.
In this technical note, the disulfide linkages of etanercept, a dimeric fusion protein, were characterized by using CID and EAD with a multi-enzyme approach (trypsin + Glu-C). The performance of CID and EAD in peptide backbone coverage for disulfides at different complexity levels was compared using the SCIEX ZenoTOF 7600 system.
Figure 1. The ZenoTOF 7600 system and Biologics Explorer software.
Key features of EAD workflow for disulfide linkage characterization
- Confident identification: Solid sequence confirmation and characterization of complex peptides with disulfide linkages were achieved using the EAD capabilities of the ZenoTOF 7600 system
- Accurate localization: Labile post-translational modifications (PTMs), such as glycosylation, were unambiguously localized on disulfide peptides
- High-quality MS/MS data: Superior fragmentation coverage was obtained, especially for long disulfide peptides that contain 2 or more disulfide bonds
- Powerful data analysis software: Superior algorithms in the Biologics Explorer software allow for comprehensive identification and spectra annotation of complex disulfide peptides
- Streamlined solution: EAD, combined with the powerful Biologics Explorer software analysis tool, provides a complete solution for challenging disulfide bond analysis
Methods
Sample preparation: Etanercept is a dimeric fusion protein that is approximately 125 kDa. It consists of 2 extracellular domains composed of the tumor necrosis factor receptor 2 (TNFR2) and the Fc part of human IgG1. The protein was denatured with 7 M guanidine-hydrochloride (GuHCl) in 100 mM Tris, pH 7.0. Then it was alkylated with 20 mM iodoacetamide. The alkylated sample underwent buffer exchange to 1M GuHCl using a 10 kDa molecular cut-off filter and then was digested overnight at 37°C with trypsin/Lys-C and Glu-C. The digestion was quenched with 1% trifluoroacetic acid.
Chromatography: LC-MS analysis was performed with an ExionLC system coupled with the ZenoTOF 7600 system (SCIEX). The protein digest was injected onto an ACQUITY CSH C18 column (2.1 × 150 mm, 1.7 μm, 130 Å, Waters) with column temperature set to 45°C. The LC method was 60 min. Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. The flow rate was set to 0.2 mL/min. The gradient used is shown in Table 1.
Table 1. LC separation gradient.
Mass spectrometry: LC-MS/MS data were acquired with data dependent acquisition (DDA) in positive ion mode on the ZenoTOF 7600 system using SCIEX OS software, version 2.1. CID and EAD MS/MS data were acquired in separate injections to allow side-by-side comparison. The same source and TOF MS parameters were used for both CID and EAD experiments. Source parameters were optimized based on the flow rate using the Turbo V ion source. Key TOF MS and MS/MS parameters used are listed in Tables 2 and 3. EAD experiments to vary the mechanism of fragmentation used to analyze specific analytes (for example, low-energy ECD or EIEIO). In this study, we used an optimized platform method with an electron KE = 7 eV to ensure efficient and comprehensive characterization of a wide range of peptides in a single injection.
Table 2. TOF MS parameters.
Data analysis: Data were processed using the peptide mapping workflow embedded in SCIEX Biologics Explorer software. The annotation of EAD data was well supported by default settings that were optimized for SCIEX EAD data in the software. The expected disulfide linkages were set as fixed modifications, as follows: Cys18-Cys31, Cys32-Cys45, Cys35-Cys53, Cys56-Cys71, Cys74-Cys88, Cys78-Cys-96, Cys98-Cys104, Cys112-Cys121, Cys115-Cys139, Cys-142- Cys157 and Cys163-Cys178 in the 2 TNFR2 regions of etanercept and Cys240-Cys240, Cys246-Cys246, Cys249- Cys249, Cys281-Cys341 and Cys387-Cys445 in the IgG1 Fc domain. The embedded O-glycan library was used to analyze peptides modified with O-glycans. All figures presented in this technical note were directly exported from Biologics Explorer software.
Table 3. MS/MS parameters for CID and EAD.
Figure 2. CID and EAD MS/MS spectra of disulfide peptide VTCVVVDVSHEDPE/CK (Cys281-Cys341).
Figure 3. CID and EAD MS/MS spectra of disulfide-bonded peptide NQVSLTCLVK/WQQGNVFSCSVMHE (Cys387-Cys445).
Figure 4. CID and EAD MS/MS spectra of disulfide-bonded peptide LPAQVAFTPYAPEPGSTCR/YYDQTAQMCCSK/VFCTK (Cys18-Cys31, Cys32-Cys45).
Identification of disulfide peptides with a single disulfide bond
The disulfide peptide VTCVVVDVSHEDPE/CK (z=4, m/z 592.2710) was subjected to CID and EAD fragmentation in 2 separate DDA runs that were performed side-by-side. In EAD MS/MS spectra, nearly 100% sequence coverage (94%) was attained with rich c/z, b/y and a/x ions. The disulfide linkages prevent each peptide from efficiently fragmenting, therefore, several peptide backbone fragments were absent from the CID MS/MS spectra, as illustrated in Figure 2.
The same pattern of EAD producing significantly more backbone fragments than CID is evident in the longer disulfide peptide NQVSLTCLVK/WQQGNVFSCSVMHE (z=4, m/z 689.0766). EAD performed particularly well in the region of the peptide near the cystine where CID was unable to produce enough fragments for peptide sequencing. The nearly 100% sequence coverage (96%) of EAD MS/MS spectra demonstrated an excellent sequencing of the disulfide peptide, as shown in Figure 3.
Identification of disulfide peptides with double disulfide bonds
The most complex disulfide linkage of etanercept lies in the TNFR region, where some peptides are connected by double or triple disulfide bonds. For example, Cys18-Cys31 and Cys32- Cys45 link 3 peptides: LPAQVAFTPYAPEPGSTCR, YYDQTAQMCCSK and VFCTK. Except for the accurate mass match of the precursor ion, CID only provided limited peptide backbone information, which hindered its ability to explicitly confirm each peptide. Only 3 b/y fragments were generated from CID for the peptide VFCTK. In contrast, EAD generated 7 c/z and b/y fragments for peptide VFCTK and almost complete sequence coverage for the other 2 peptides, LPAQVAFTPYAPEPGSTCR and YYDQTAQMCCSK (Figure 4). This comparison demonstrates that EAD can sequence disulfide peptides successfully, even when 4 cysteines are connected by 2 disulfide links.
Identification of disulfide peptides with additional O-glycosylation
Etanercept is well-known for its complex glycosylation profiles. Eleven possible O-linked glycosylation sites (serine/threonine) are present in the hinge region, which makes analysis difficult. Here, the peptide pair, THTCPPCPAPE/THTCPPCPAPE linked by 2 disulfide bonds with an additional O-glycosylation modification, was confidently identified by EAD. Additionally, the presence of a c3 ion with an O-glycan (Hex1HexNAc1NeuAc1) and a c2 ion without an O-glycan indicated that the Oglycosylation site was located at the third, but not first, threonine residue, as shown in Figure 5. The presence of another c3 ion without an O-glycan, combined with the precursor accurate mass match, demonstrated partial glycosylation occupancy. In summary, EAD provided the exceptional capability for complete disulfide linkage mapping of etanercept for challenging disulfide peptides with additional O-glycan modifications.
Figure 5. CID and EAD MS/MS spectra of disulfide-bonded peptides THT*CPPCPAPE/THT*CPPCPAPE (Cys246-Cys246, Cys249-249). Oglycosylation sites are labeled with a star.
Conclusion
- EAD provides superior sequence and disulfide linkage confirmation, especially for long disulfide peptides, compared to traditional fragmentation mechanisms
- For peptides that contain 2 or more disulfide bonds, EAD generated rich fragments that permitted reliable sequencing of every peptide involved in the disulfides and demonstrated superior fragmentation coverage compared to CID
- EAD preserves the labile O-glycan modification, which enables site-specific characterization of O-glycosylated disulfide-bonded peptides
- EAD provides excellent backbone sequencing for disulfide peptides, while simultaneously enabling unambiguous localization of the O-glycosylation site
References
- Baba T et al. (2015) Electron capture dissociation in a branched radio-frequency ion trap, Anal Chem, 87,785−792.
- Baba T et al. (2021) Dissociation of biomolecules by an intense low-energy electron beam in a high sensitivity time-offlight mass spectrometer. J. Am. Soc. Mass Spectrom. 32(8):1964-1975.2.
- Confirmation of disulfide linkages in adalimumab using electron activated dissociation, SCIEX technical note, RUOMKT-02-12913-B
 Click to enlarge
Click to enlarge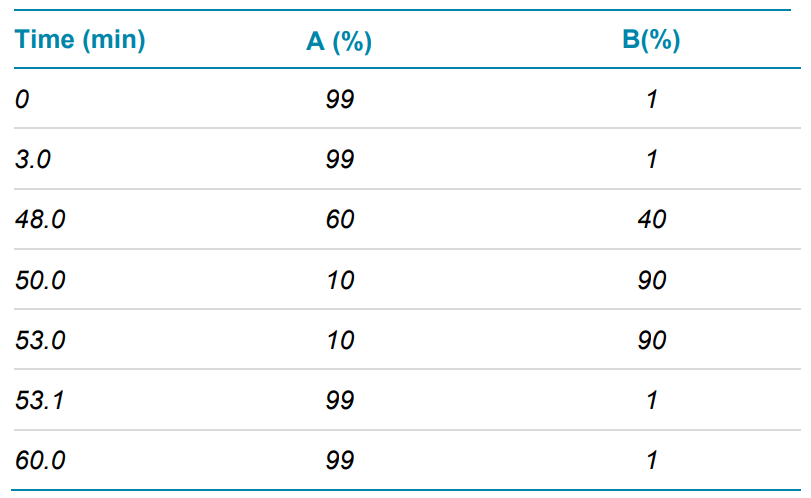 Click to enlarge
Click to enlarge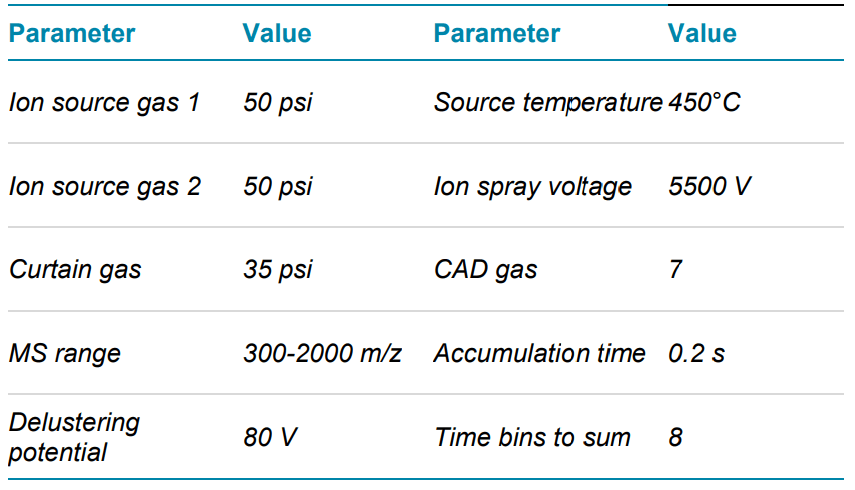 Click to enlarge
Click to enlarge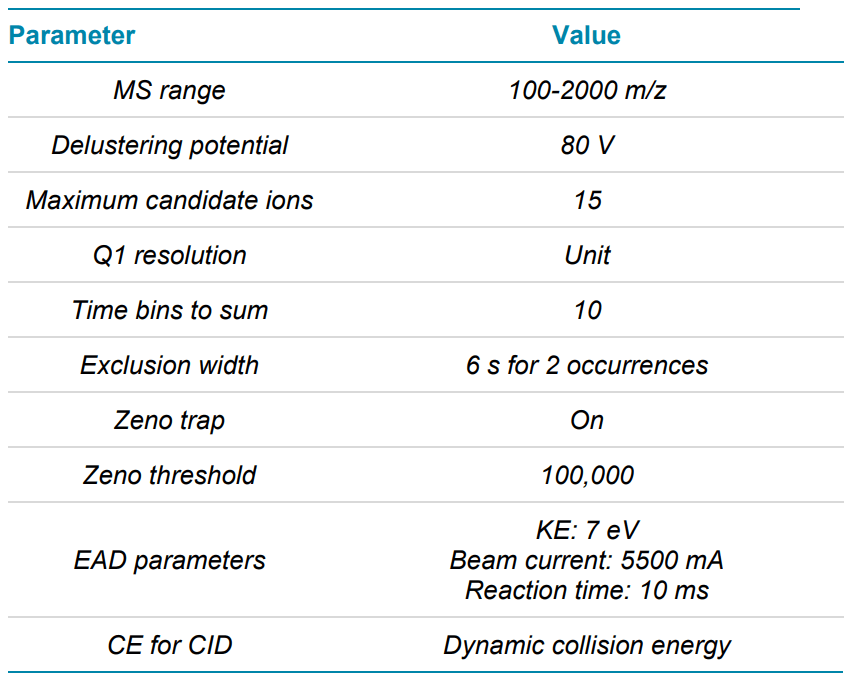 Click to enlarge
Click to enlarge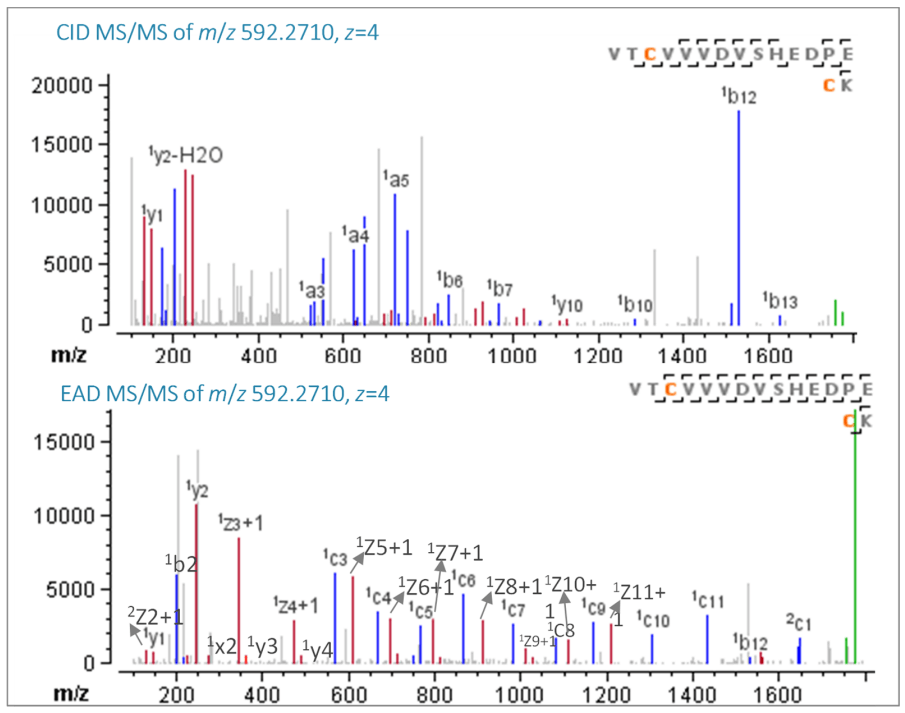 Click to enlarge
Click to enlarge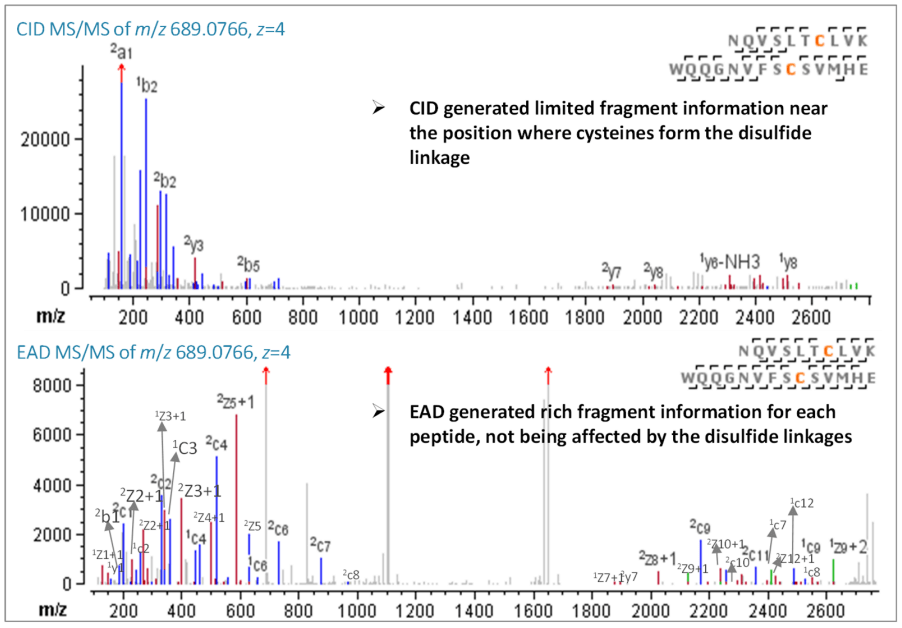 Click to enlarge
Click to enlarge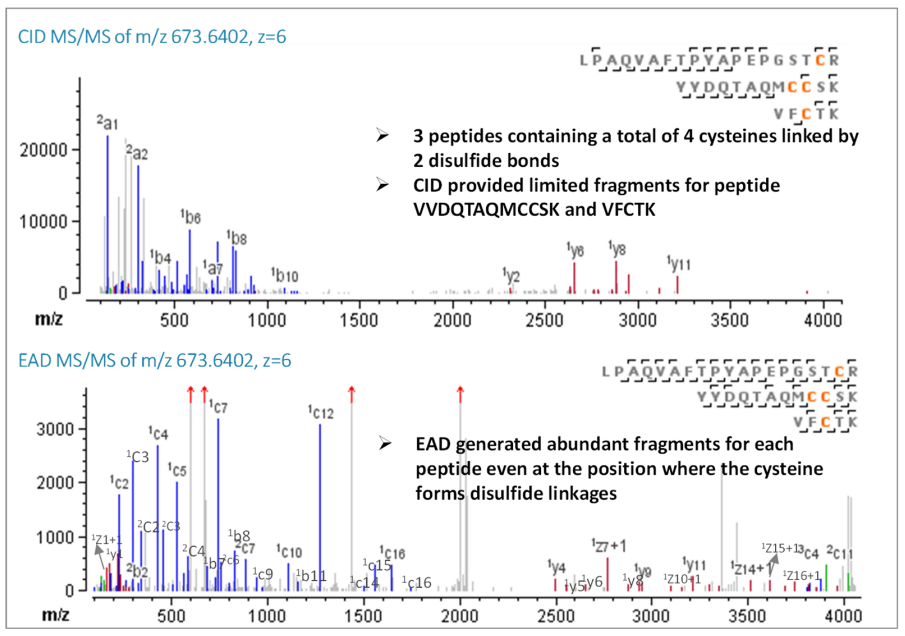 Click to enlarge
Click to enlarge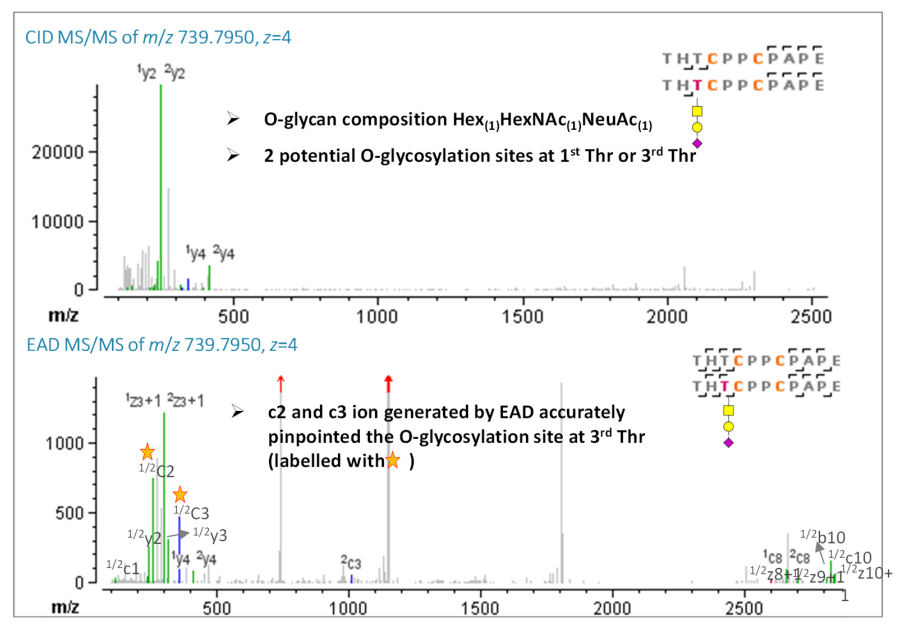 Click to enlarge
Click to enlarge