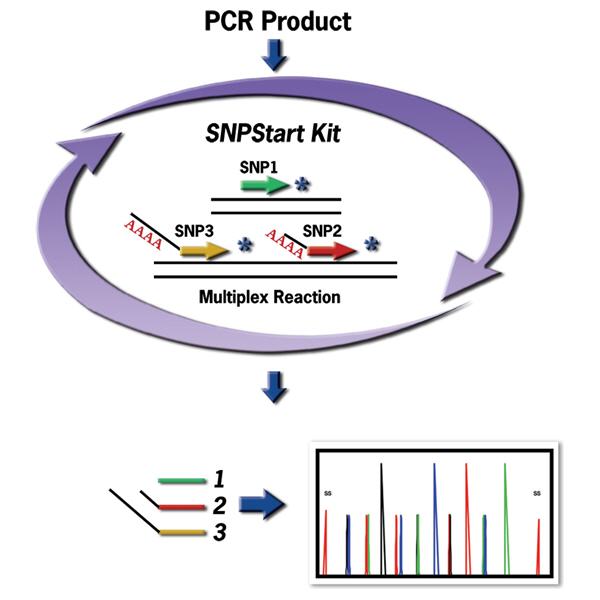
SNP analysis is routinely used for linkage, association studies, pharmacogenetics, forensics, etc. Similarly, mutation screening requires detection of single base changes, as well as analysis of insertions, deletions and translocation among other molecular events. Single-Base Extension (SBE) technology enables SNP analysis by extending an unlabeled primer when hybridized to a target site with a fluorescent labeled terminator. The GenomeLab SNPStart Primer Extension Kit provides a quick and robust ready-to-use reagent for single or multiplex identification of DNA sequence variations. The master mix is optimized to generate accurate as well as well-balanced allele signals in a SNP reaction.
Template in the form of PCR* product can be used for SNP reactions. These products should be free of non-specific products and other artifacts. Specificity can be increased by designing PCR primers with high annealing temperature. The desired lengths of the PCR products are between 150-1000 bps, where smaller PCR products are preferred since they have a less likelihood of nonspecific binding to other products. For multiplex PCR situation, the product lengths can be designed so that they do not overlap by less than 30 bps.
There are two choices in designing the interrogation primers, either on the forward strand or the reverse strand. These interrogation primers are then labeled using complimentary bases during primer extension SNP reaction. SNP analysis occurs during capillary electrophoresis fragment separation on the GenomeLab GeXP Genetic Analysis System.
*The PCR process is covered by patents owned by Roche Molecular Systems, Inc. and F. Hoffmann-La Roche, Ltd.