Characterization of an antibody-drug-conjugate (ADC) using electron activated dissociation (EAD)
Featuring the SCIEX ZenoTOF 7600 system with EAD and Protein Metrics Inc. software
Zoe Zhang1 , Takashi Baba2 , Pavel Ryumin2 , Bill Loyd2 , Jason Causon2 , Kerstin Pohl1
1SCIEX, US; 2SCIEX, Canada
Abstract
Here, the characterization of lysine-linked drugs in an ADC sample is demonstrated. A bottom-up approach was chosen to determine the position of the conjugation. Alternative fragmentation using electron activated dissociation (EAD)1,2 was leveraged for detailed structural information on the drug and linker, along with obtaining peptide backbone information.
With the development in protein engineering, antibodies and their related derivatives become the fastest growing class of therapeutics.3 ADCs are one of those new modalities. ADCs a are often composed of a 150 kDa monoclonal antibody (mAb) covalently coupled with cytotoxic payloads, or other types of drugs, through synthetic linkers.4 ADCs show a more complex structure and heterogeneity compared to unconjugated proteins, since the addition of a variable number of payload and linkers can significantly enhance the number of proteoforms.5 To ensure drug safety and efficacy, an in-depth characterization of ADCs is essential during their development. This includes not only the identification and the localization of post-translational modifications (PTMs) on the mAb, but also a verification of the drug conjugation. Mass spectrometry (MS) has become the most widely employed method for ADC characterization, owing to the rapid advancement of MS technologies. Intact mass analysis is the platform method utilized to determine drug-to-antibody ratio (DAR), while deep characterization of the sites of conjugation usually relies on bottom-up approaches. Most widely-adopted, collision-induced dissociation (CID) is able to provide amino acid sequence confirmation, but the harsh fragmentation technique also breaks the payload into small pieces. The highly complex spectra derived from such an approach can be very difficult to interpret. Alternative fragmentation can provide further insights into such complex samples, but previous techniques suffered from long reaction times, low sensitivity and lack of reproducibility.
A new, highly reproducible fragmentation type based on EAD1,2 was used to analyze the conjugated peptides from a commercial ADC. The data were acquired with an untargeted 10 Hz rapid data-dependent acquisition (DDA) method and interpreted with Protein Metrics Inc. software. With this workflow, regular and advanced characterization leveraging EAD-based fragmentation is achievable in one injection, enabling a streamlined characterization accessible to every user-level.
Key features of the SCIEX ZenoTOF 7600 system
- New depths of peptide mapping analysis: EAD with fast DDA enables alternative fragmentation for routine, in-depth analysis of next generation protein therapeutics and standard mAbs
- Higher levels of structural information: Changing the mechanism of fragmentation by tuning the electron energy may provide a higher level of structural information
- Higher MS/MS sensitivity: Increased detection of fragments (5 to 10 fold) using the Zeno trap enables higher confidence in data assignment
- High reproducibility: Reproducible fragmentation with EAD for singly, doubly, and multiply charged ions enables analysis of more precursors than other alternative and low reproducibility fragmentation techniques
- Streamlined and easy-to-use: Fully automated data acquisition in DDA mode using EAD with SCIEX OS software, and automated data interpretation with Byos software (Protein Metrics Inc.) simplifies the entire user experience
Figure 1. The SCIEX ZenoTOF 7600 system.
 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge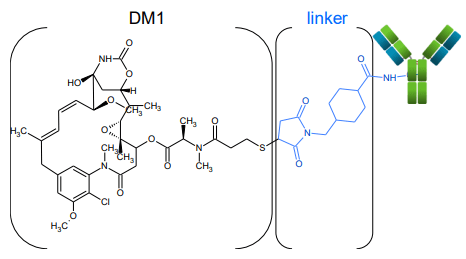 Click to enlarge
Click to enlarge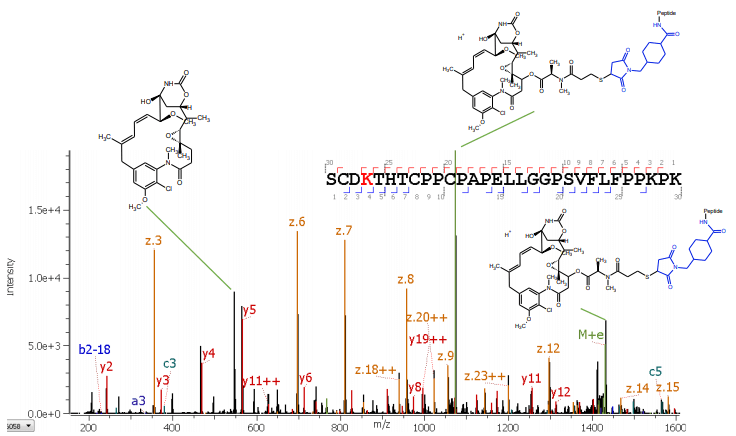 Click to enlarge
Click to enlarge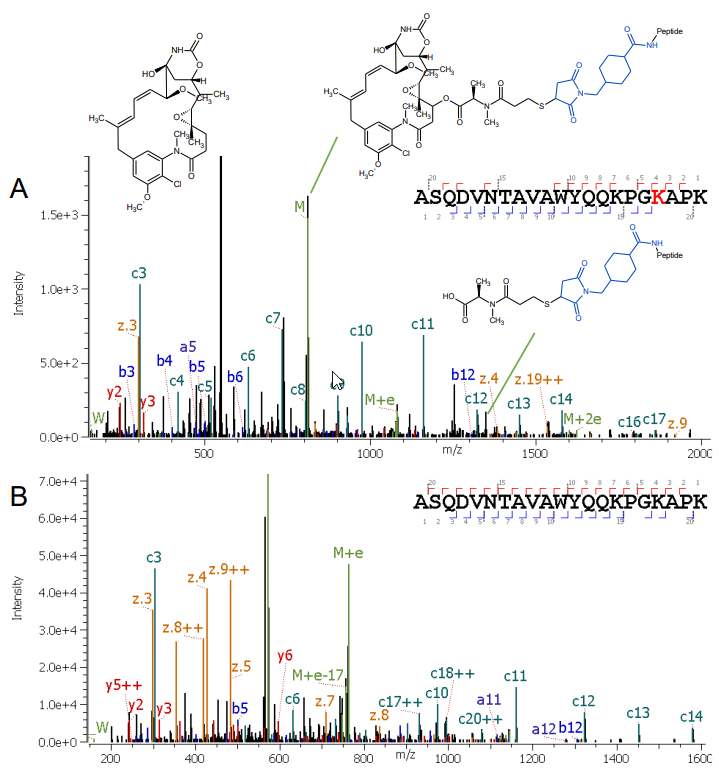 Click to enlarge
Click to enlarge