Comprehensive peptide mapping of biopharmaceuticals utilizing electron activated dissociation (EAD)
Featuring the SCIEX ZenoTOF 7600 system with EAD and Protein Metrics software
Zoe Zhang1, Kerstin Pohl1, Takashi Baba2, Pavel Rumin2, Bill Loyd2, Jason Causon2 and Elliott Jones1
1SCIEX, US; 2SCIEX, Canada
Abstract
The automated characterization of challenging post-translational modifications (PTMs), the confirmation of amino acid (AA) isomers and a streamlined, simplified disulfide mapping approach will be presented in this work. An unprecedented in-depth characterization can be routinely achieved using electron activated dissociation (EAD)1,2 as part of a fully automated data-dependent acquisition (DDA) workflow that includes processing with Protein Metrics software.
As next generation biologics become more complex and sophisticated in terms of structure, the requirements of analytical tools for characterization and quality assessment are consequently increasing. Traditionally used collision-induced dissociation (CID) has a number of limitations in PTM localization. It is limited in its ability to determine peptide side chains and in its disulfide mapping capabilities. Alternative fragmentation, however, struggled with sensitivity, reproducibility, and acquisition speed. These fundamental limitations have restrained the widespread adoption of alternative fragmentation as the primary technique for peptide mapping.
Developed to address the rigorous analytical challenges of biopharmaceuticals, the ZenoTOF 7600 system (Figure 1) is uniquely capable of facilitating EAD at speeds up to 20 Hz and offers sensitivity enhancements using the patented Zeno trap. In addition, quantitative reproducibility is accomplished through a reagent-free fragmentation technique. In-depth peptide mapping of new and complex molecules, as well as standard monoclonal antibodies (mAb), can be achieved with the system in a fully automated fashion. Increased MS/MS sequence coverage, PTM characterization and AA isomer elucidation can be accomplished with this newly designed type of mass spectrometer.
Key features of the SCIEX ZenoTOF 7600 system
- New depths of peptide mapping analysis: EAD with fast DDA enables alternative fragmentation for routine, in-depth analysis of next generation protein therapeutics and standard mAbs
- Higher levels of structural information: Changing the mechanism of fragmentation by tuning the electron energy may provide a higher level of structural information, particularly suited for glycopeptides, AA isomer differentiation and disulfide mapping
- Higher MS/MS sensitivity: Increased detection of fragments (5 to 10 fold) using the Zeno trap enables higher confidence in data assignment
- High reproducibility: Reproducible fragmentation with EAD for singly, doubly, and multiply charged ions enables analysis of more precursors than other alternative and low reproducibility fragmentation techniques
- Streamlined and easy-to-use: Fully automated data acquisition in DDA mode using EAD with SCIEX OS software, and automated data interpretation with Byos software (Protein Metrics Inc.) simplifies the entire user experience
Figure 1. The SCIEX ZenoTOF 7600 system. The system is a new type of mass spectrometer combining ease-of-use with excellent data quality and remarkable versatility.
 Click to enlarge
Click to enlarge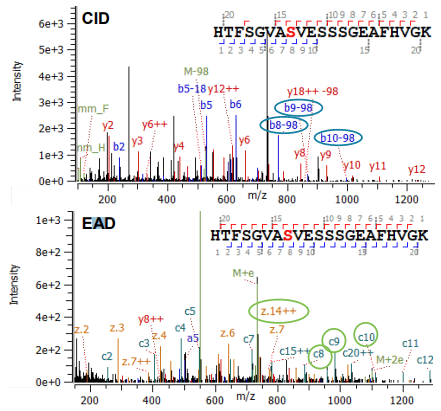 Click to enlarge
Click to enlarge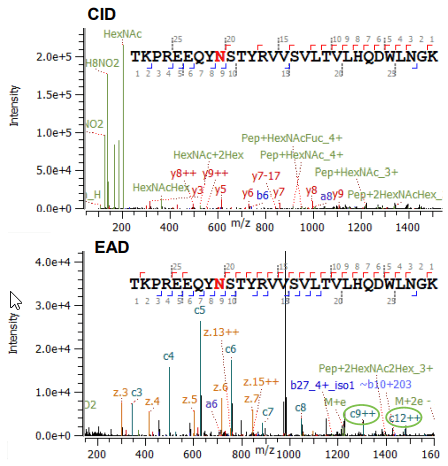 Click to enlarge
Click to enlarge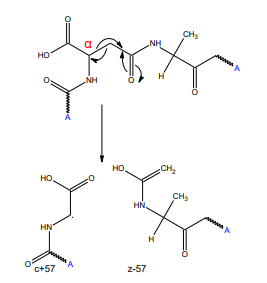 Click to enlarge
Click to enlarge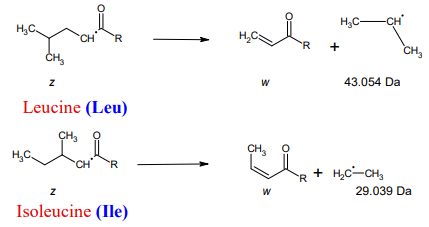 Click to enlarge
Click to enlarge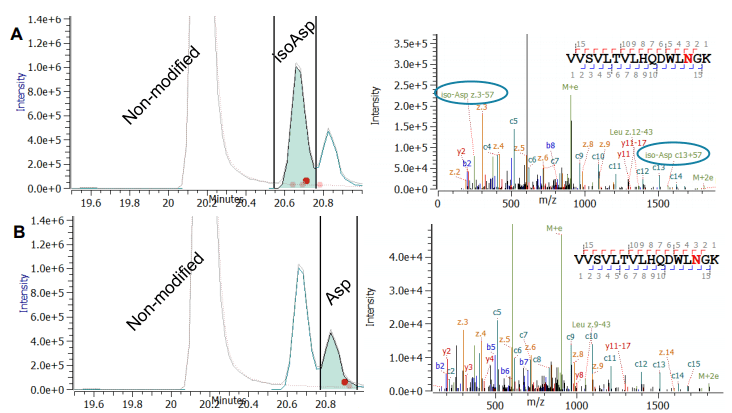 Click to enlarge
Click to enlarge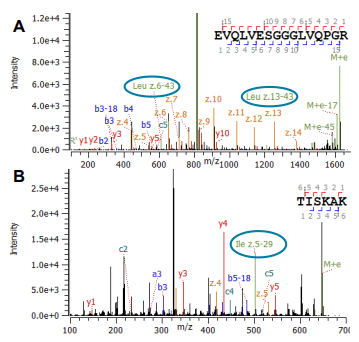 Click to enlarge
Click to enlarge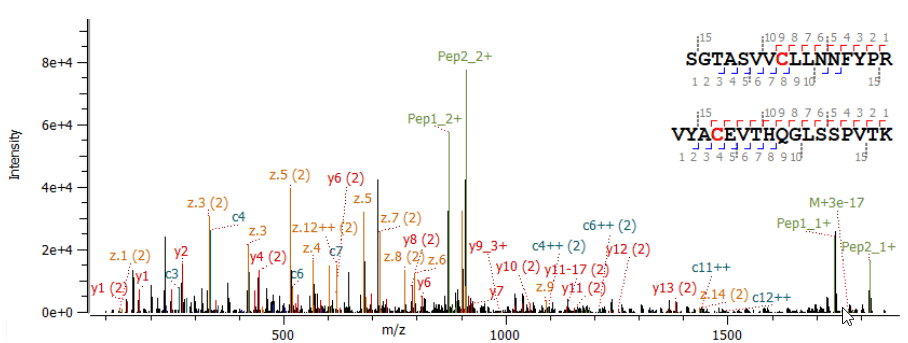 Click to enlarge
Click to enlarge