Abstract
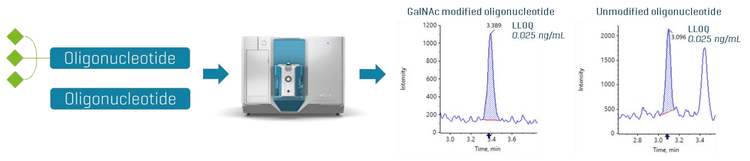
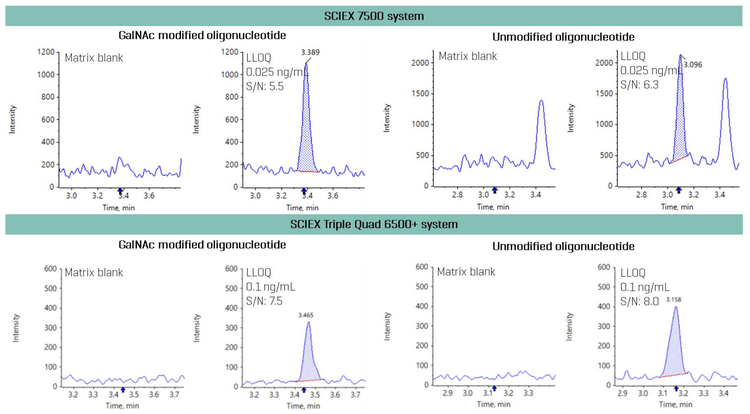
This technical note demonstrates a sensitive method for the quantitation of GalNAc modified and unmodified oligonucleotides using the SCIEX 7500 system. The lower limit of quantitation (LLOQ) of 0.025 ng/mL was achieved in plasma samples (Figure 1). A 4-fold improvement in LLOQ was achieved on the SCIEX 7500 system compared to SCIEX Triple Quad 6500+ system.
Oligonucleotide therapeutics are rapidly gaining attention as their potency improves and delivery challenges are addressed. Modalities such as antisense oligonucleotides (ASOs) and small interfering RNAs (siRNAs) are becoming more critical due to their high specificity and ability to reach formerly untreatable targets. Sensitive and robust methods for quantitative analysis of oligonucleotides are needed to support the development of these therapeutics. Hybridization methods can achieve sensitive detection limits. However, this type of assay may be unable to differentiate oligonucleotides from impurities and metabolites.
LC-MS/MS can provide excellent selectivity, high sensitivity and the ability to analyze multiple oligonucleotides in a single assay. Here, a sensitive triple quadrupole mass spectrometer was applied for the analysis of complex oligonucleotides in biological matrix.
Key benefits for analysis of GalNAC modified and unmodified oligonucleotides in plasma using the SCIEX 7500 system
- Sub ng/mL level of quantitation: Achieve 0.025 ng/mL LLOQ for quantitation of GalNAc modified and unmodified oligonucleotides in plasma
- Increased sensitivity levels: Reach a 4-fold improvement in LLOQ using SCIEX 7500 system compared to SCIEX Triple Quad 6500+ system
- Robust analytical performance: Achieve accurate quantitative performance with %CV <14% at all concentration levels across a linear dynamic range (LDR) of 3.6 orders of magnitude
- Enhanced sensitivity unlocked: Improved front-end technology with the D Jet ion guide, OptiFlow Pro ion source and E Lens probe for enhanced ion generation, capture and transmission, enabling users to reach their desired quantitative sensitivity1
- Streamlined data management: SCIEX OS software, a 21 CFR Part 11-compliant platform, simplifies data acquisition and processing
Introduction
Antisense oligonucleotides (ASOs) effectively reduce disease-causing mRNAs by utilizing the body's natural RNA interference (RNAi) pathway. Consequently, scientists are actively developing these nucleic acid-based therapeutics for a wide range of both rare and common diseases.2 One promising approach involves enhancing delivery by attaching trivalent GalNAc, thereby improving both the potency and duration of action of these therapies.3 The growing interest in GalNAc-modified ASOs requires the development of appropriate bioanalytical methods to understand their pharmacokinetic profiles and metabolism. In this study, GalNAc-modified and unmodified oligonucleotides were measured in plasma using a SCIEX Triple Quad 6500+ system and SCIEX 7500 system in order to compare their direct performance using the LC-MS/MS method for the simultaneous determination of GalNAc-conjugated 18 mer single-stranded oligonucleotide (GalNAc-ASO1) and unconjugated form (ASO1) in the same rat plasma samples.
Methods
Sample preparation: Calibration standards samples (100 µL) underwent protein denaturation with 150 Qiagen RLT lysis buffer containing 1% (v/v) beta-mercaptoethanol. This was performed after the addition of 20 µL of 500 ng/mL single strained 18 mer ASO labeled with 34S atoms internal standard (34S-ASO-IS) into 1:4 (v/v) methanol/ water solution. After vigorous mixing for 20 min at 1700 rpm, a 300 µL aliquot of 50mM ammonium acetate water solution at pH 5.5 was added. The solution was then mixed for 15 min at 1600 rpm. Then a clean-up step was performed by means of solid-phase-extraction 96-well cartridges (Phenomenex STRATA X-AW, 10 mg sorbent). The Phenomenex Strata X-AW 96 well-plate (DWP) was primed with 300 μL of methanol followed by 300 μL of 50mM ammonium acetate at pH 5.5 solution. The plate was washed with 500 μL of 50mM ammonium acetate at pH 5.5 and 500 μL of 50:50 (v/v) 50mM ammonium acetate at pH 5.5/methanol. The analytes and internal standard were eluted using 300 μL (2 x 150 μL) 60:40 (v/v) 100mM ammonium bicarbonate at pH 10/acetonitrile. After elution and evaporation to dryness (60 - 75 min under a stream of nitrogen at +40°C), the samples were reconstituted in 150 µL 95 : 5 : 1 : 0.3 (v/v/v/v) water/acetonitrile/hexafluoroisopropanol/diisopropylethylamine. After vortexing for 10 min at 1500 rpm, the samples were placed under centrifugation for 10 min at 5600 rpm.
Chromatography: Analytical separation was performed on the Nexera XS inert HPLC system using an ACQUITY Premier Oligonucleotide BEH C18 (2.1 × 50 mm, 1.7 μm, 130 Å) column at a flow rate of 0.4 mL/min. Mobile phase A was 95:5 (v/v) water/acetonitrile in 1% HFIP and 0.3% DIPEA and mobile phase B was 60:40 (v/v) water/acetonitrile in 1% HFIP and 0.3% DIPEA. The column temperature was set to 60°C. The gradient conditions used are summarized in Table 1. A 20 μL aliquot of the sample was injected for LC-MS/MS analysis.
Data processing: Data collection and analysis were performed using SCIEX OS software, version 3.3.1.
The same rat plasma extracts from the calibration curves were analyzed for GalNAc modified and unmodified oligonucleotides at concentrations ranging from 0.025 to 100 ng/mL using both the SCIEX 7500 system and the SCIEX Triple Quad 6500+ system. The chromatographic conditions were equivalent on both platforms, to ensure a proper comparable assessment. Each calibration standard was analyzed in 5 replicates to evaluate reproducibility. Analytical performance on both the SCIEX 7500 system and the SCIEX Triple Quad 6500+ system were evaluated based on the requirement that the accuracy of the calculated mean should be between 80% and 120% at the LLOQ and between 85% and 115% at higher concentrations. The %CV of the calculated mean of the concentration should be below 20% at the LLOQ and below 15% at all higher concentrations.6
Quantitative performance on the SCIEX 7500 system
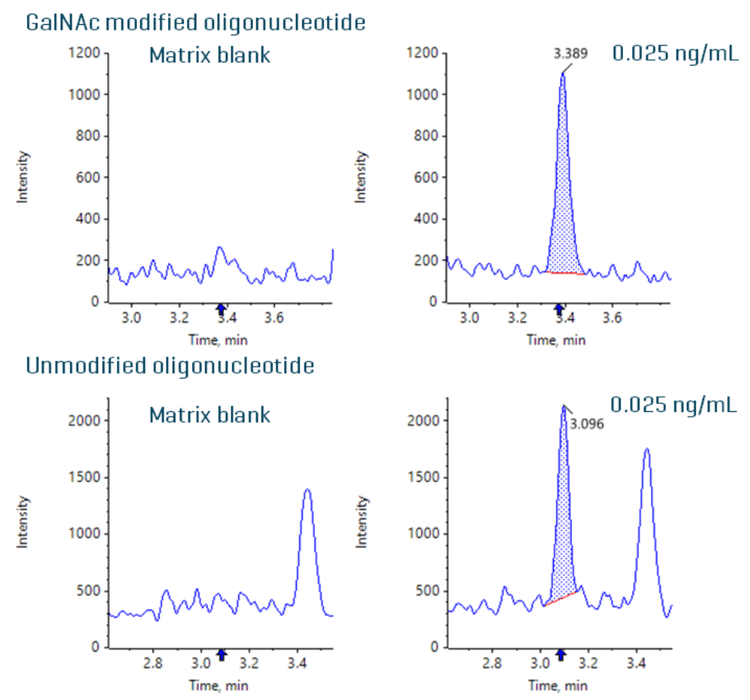
GalNAc modified and unmodified oligonucleotides were quantified in plasma at an LLOQ of 0.025 ng/mL. No interferences were observed in the matrix blank for the GalNAc modified and unmodified oligonucleotides (Figure 2).
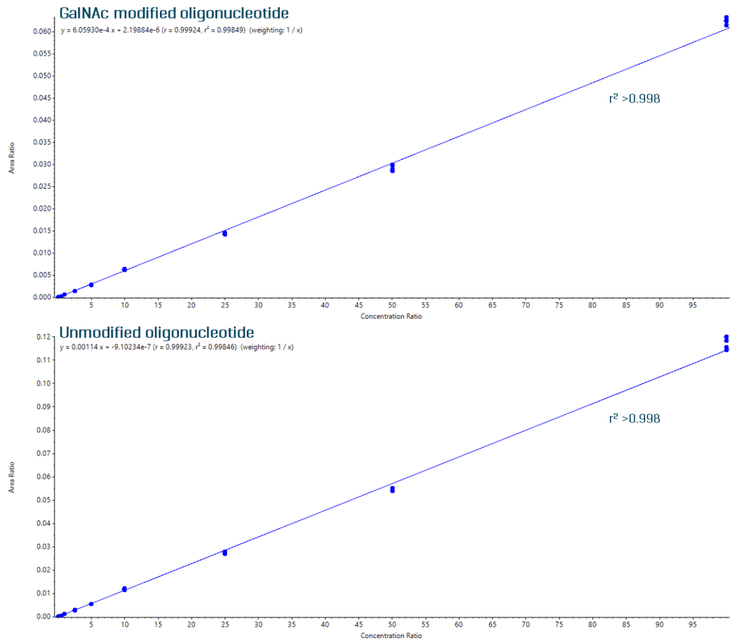
Linearity was achieved across concentrations ranging from 0.025 to 100 ng/mL, with a coefficient of determination (r2) of >0.998 for both analytes (Figure 3). Both analytes achieved a linear dynamic range (LDR) of 3.6 orders of magnitude.
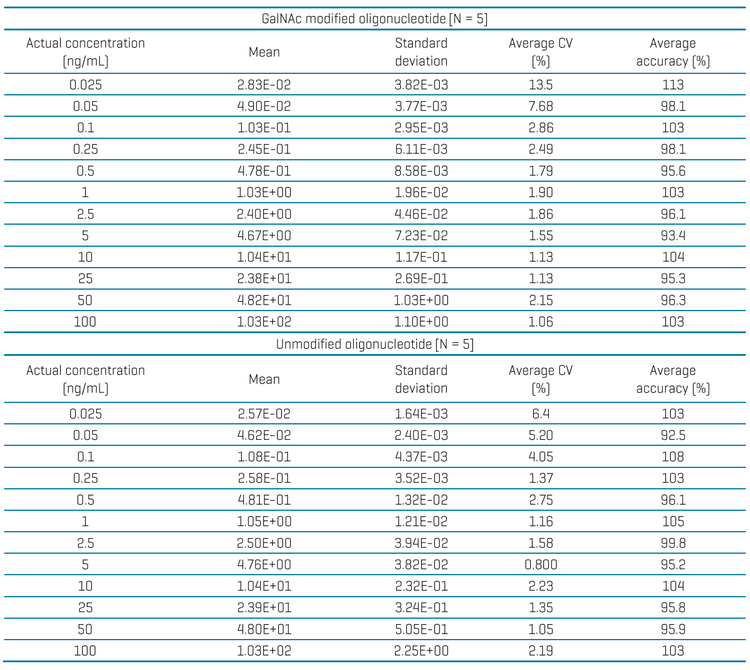
For this assay, accuracy was between 92.5% and 113% and %CV was <14% for GalNAc modified and unmodified oligonucleotides in plasma (Table 4). Calculated percent accuracy and %CV values were within the acceptance criteria at each concentration level.
After injecting the upper limit of quantitation (ULOQ, 100 ng/mL), carryover was >20% of the LLOQ for both the GalNAc modified and unmodified oligonucleotides in the blank. Therefore, the useable ULOQ could be lower. Further work on reducing the carryover will be required to fully utilize the demonstrated dynamic range.
Comparing quantitative performance with SCIEX Triple Quad 6500+ system
Quantitation of GalNAc modified and unmodified ASOs was also evaluated on the SCIEX Triple Quad 6500+ system. A 4-fold improvement in LLOQ was achieved on the SCIEX 7500 system compared to the SCIEX Triple Quad 6500+ system (Figure 4). No interferences were observed in the matrix blank for the GalNAc modified and unmodified oligonucleotides on either MS platform.
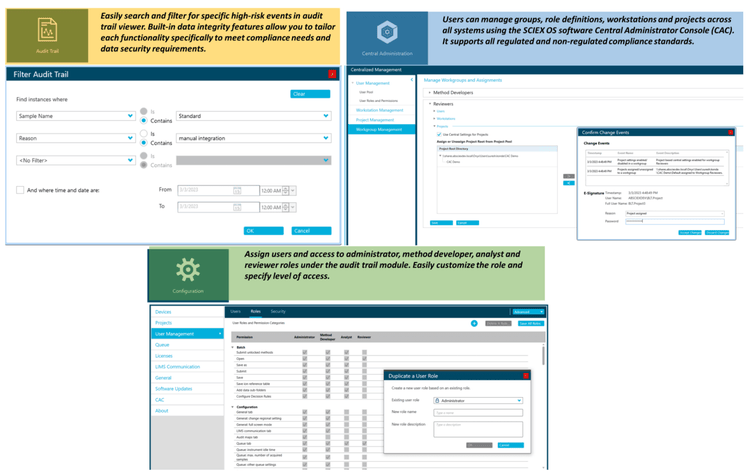
Compliance-ready SCIEX OS software
Equivalent SCIEX OS software capabilities for regulated bioanalysis can be executed on the SCIEX 7500 system, ensuring high fidelity when performing method transfers while retaining critical compliance features.
SCIEX OS software is a closed system and requires records and signatures to be stored electronically, meeting the regulations outlined by 21 CFR Part 11. SCIEX OS software can open raw data files from any visible storage location within a closed network by using designated processing workstations.
Figure 5 illustrates the features of SCIEX OS software used to monitor the audit trail, acquire and process data, and configure user access. The audit trail feature enables users to audit critical user actions and locks in data integrity. The Central Administrator Console (CAC) feature allows users to centralize acquisition and processing using a single platform to maximize efficiency for multi-instrument laboratories, independent of compliance standards. The configuration module allows users to assign roles and access as the administrator, method developer, analyst, and reviewer.
Conclusions
- An LLOQ of 0.025 ng/mL was achieved for quantitation of GalNAc modified and unmodified oligonucleotides in plasma
- A 4-fold improvement in LLOQ was observed using the SCIEX 7500 system compared to the SCIEX Triple Quad 6500+ system
- Linearity was achieved at concentrations ranging from 0.025 ng/mL to 100 ng/mL with an r2 >0.998 for both analytes
- Comparable quantitative performance was demonstrated with accurate and highly reproducible (%CV <14%) results on the SCIEX 7500 system
- Sensitivity was achieved on the SCIEX 7500 system with an improved front-end technology for better ion generation, capture and transmission
- Data management and compliance-readiness (21 CFR Part 11) features using SCIEX OS software to support regulated bioanalysis was retained on the SCIEX 7500 system
References
- SCIEX 7500 system. SCIEX brochure, RUO-MKT-03-11712-A.
- E., Martin; M., Muthiah. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023; 51:2529–2573.
- Y., Mikung, et. al. Evaluating the oral delivery of GalNAc-conjugated siRNAs in rodents and non-human primates. Nucleic Acids Res. 2024; 52(10): 5423-5437.


