Abstract
The poly(A) tail is an essential part of messenger RNA (mRNA). For mRNA-based therapeutics and vaccines, the length of the poly(A) tail is a critical quality attribute (CQA) since it directly affects the stability and translation efficiency of the mRNA. In this technical note, we describe a workflow for mRNA poly(A) tail analysis by CGE-UV using the ssDNA 100-R kit on the PA 800 Plus Pharmaceutical Analysis system. Single-nucleotide resolution was obtained over an extended size range of 9 to 156 nucleotides (nt), enabling accurate poly(A) tail length determination and peak quantitation with excellent assay repeatability. The simple sample preparation and ready-to-use ssDNA 100-R kit on a 21 CFR Part 11 compliance-ready platform make this a potential workflow for development of a QC release test.
Key features of mRNA poly(A) tail analysis by CGEUV on the PA 800 Plus system
- Exceptional resolving power: Achieve single-nucleotide resolution critical for accurate poly(A) length determination and peak quantitation of each poly(A) tail species
- Extended size coverage: Sustain single-nucleotide resolution in the size range of 9 to 156 nt
- Excellent assay repeatability: Obtain consistent poly(A) tail profiles in multiple injections
- QC release-friendly: Simple sample preparation for direct measurement of poly(A) tail length, ready-to-use ssDNA 100-R kit on a 21 CFR Part 11 compliance-ready platform make this workflow amenable to QC testing
- Streamlined workflow: Achieve direct measurement of poly(A) tail length and distribution with streamlined sample preparation, using the ready-to-use ssDNA 100-R kit on a 21 CFR Part 11 compliance-ready platform. This method has the potential to be developed for QC testing
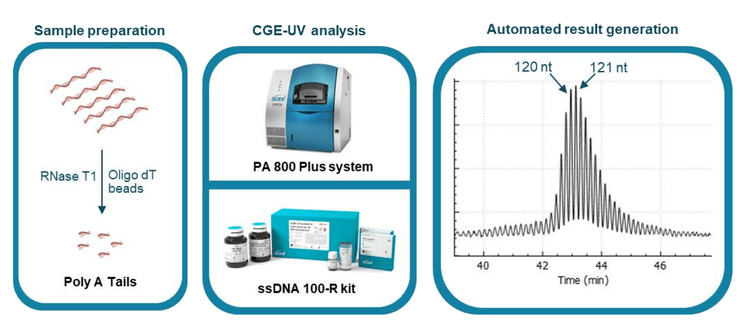
Introduction
Since the approval of mRNA vaccines against SARS-CoV-2virus, in vitro transcribed (IVT) mRNA has gained popularity in vaccine development, protein replacement therapies, and cancer immunotherapies. The poly(A) tail length is a CQA for mRNAbased therapeutics. One challenge in studying poly(A) tails is that they are difficult to sequence and accurately measure. Various analytical methods have been developed to characterize this CQA. Ion-pair reversed-phase liquid chromatography (IPRP-LC) allows quantification of the percentage of mRNA containing a poly(A) tail but lacks the ability to determine the specific length of the poly(A) tail. Next-generation sequencing (NGS) can achieve single-nucleotide resolution in separating poly(A) tail length. However, its implementation for release testing of manufactured mRNA poses complexity and requires powerful bioinformatics software tools. In this technical note, single-nucleotide resolution was achieved across an extended size range from 9 to 156 nt using a CGE-UV workflow, allowing for precise determination of poly(A) tail length and accurate quantitation of peaks with outstanding assay repeatability.
Methods
Materials: The ssDNA 100-R kit (P/N 477480) containing DNA Capillary, ssDNA 100-R Gel (lyophilized), Tris-Borate buffer, 7M Urea, and pd(A) 40-60 Test Mix; Universal Vials (P/N A62251), Universal Vial Caps (P/N A62250), PCR Microvials (P/N 144709), CE Grade Water (P/N C48034), and NanoVial (P/N 5043467) were from SCIEX (Framingham, MA). Nuclease-free water (NFW, P/N AM9932), RNase T1 (1000 U/μL, P/N EN0541), Dynabeads Oligo (dT)25 (P/N 61002), and Zeba Spin Desalting columns (7K MWCO, 75 μL, P/N 89877) were from Thermo Fisher Scientific (Waltham, MA). The 10x RNase H Reaction Buffer (Component #B0297SVIAL) of the RNase H kit (P/N M0297L) was from New England Biolabs (Ipswich, MA). The firefly luciferase (FLuc) mRNA (P/N: L-7602) was from TriLink BioTechnologies (San Diego, CA). The FLuc mRNA contains a 5’ Cap and a 3’ poly A tail, mimicking an mRNA drug substance. The 120 nucleotide (nt) polyA size marker was custom-synthesized by Integrated DNA Technologies (Coralville, IA).1 The Agencourt SPRIPlate 96S Super Magnet (P/N A32782) was from Beckman Coulter Life Sciences (Indianapolis, IN). The 0.2 µm (P/N: 4612) and the 0.45 µm (P/N 4654) syringe filters were from PALL (Port Washington, NY). The LABQUAKE rotator (model 400100) was from Barnstead International (Dubuque, IA).
Instruments and software: A PA 800 Plus Pharmaceutical Analysis System (P/N A66528) equipped with an ultraviolet (UV) detector was from SCIEX. The UV wavelength used was 254 nm. Data acquisition and analysis were performed using 32 Karat software version 10.
Tris-Borate-Urea (TBU) buffer preparation: This step must be done one or a few days before running samples. To rehydrate the buffer, 135 ml of 0.2 µm filtered deionized water was added to the bottle containing the dry Tris-Borate from the ssDNA 100-R kit. The buffer solution was mixed with a clean magnetic stirring bar for about 20–30 minutes until boric acid was completely dissolved. The dry 7M Urea was then slowly added to the Tris-Borate buffer while stirring the solution. The urea was completely dissolved after about 2 hours and the solution became clear. The reconstituted Tris-Borate-Urea buffer should be good for 30 days if stored at 2°C to 8°C after preparation. The entire bottle of buffer should be brought to ambient temperature before use. The required volume to be used for the day was removed and filtered through a 0.2 μm disposable syringe filter into a clean container.
ssDNA 100-R Gel buffer reconstitution: Five milliliters of freshly filtered TBU buffer was added to the bottle with lyophilized gel. The gel bottle was then placed onto a rotator in a cold room (2°C to 8°C) for 72 hours with gentle rotation or stirred at room temperature for 5 to 6 hours. The prepared gel buffer can be used up to 30 days after preparation if stored at 2°C to 8°C. The gel buffer was brought to ambient temperature and filtered through a 0.45 μm disposable syringe filter before use.
Cartridge assembly: A DNA capillary (PN 477477) from ssDNA 100-R kit was installed according to the instructions in the ssDNA 100-R kit application guide.2 The total capillary length was 30.2 cm, with 20 cm as the length to the detection window. A 100 x 200 μm aperture was used for better resolution when using a UV detector. Since the inner wall of the DNA capillary is coated, the cartridge assembly should be carried out promptly. Excessive exposure to air may damage the inner coating and cause clogging. The capillary ends must be immersed in liquid (water or buffer) when the cartridge assembly is complete to prevent the coating from drying out.
Preparation of buffer trays and sample trays: Vial positions for buffer trays are indicated in Figure 2A. Each CE Water vial was 3 filled with 1.5 mL CE Grade Water. The waste vial was filled with 1.0 mL CE Grade Water. The 100-R Gel vial was filled with 1.5 mL ssDNA 100-R Gel buffer, and the TBU Buffer vials were filled with 1.5 mL TBU buffer.
Instrument setup: The Initial conditions and UV detector initial conditions were set up as indicated in the ssDNA 100-R kit application guide.2 The same setup was used for all methods: the new capillary-conditioning method, the gel-filling method, the separation method and the shutdown method. The time program settings for the capillary-conditioning method and for the gel-filling method are provided in the ssDNA 100-R kit application guide.2 Time program settings for the separation method are shown in Figure 2B.
Poly(A) tail analysis workflow by CGE-UV with single nucleotide resolution
The workflow of Poly(A) tail analysis by CGE-UV is illustrated in Figure 1. The mRNA sample is digested with RNase T1 first. The poly(A) tails released are then purified with oligo dT conjugated magnetic beads and separated on the PA 800 Plus system using the ssDNA 100-R kit that contains a replaceable gel buffer and a coated capillary for enhanced reproducibility. The electropherogram in the right panel demonstrates single-nucleotide resolution in a typical poly(A) tail profile obtained with an mRNA sample.
Poly(A) tail length determination
To determine the poly(A) tail length, the poly(A) tails released from the TriLink FLuc mRNA sample were mixed with a 120 nt size marker before CE separation on the PA 800 Plus. As a control, the poly(A) tail samples were also analyzed alone. Results obtained with the poly(A) tail sample and the spiked-in 120 nt size marker are shown in the upper panel of Figure 4, while the results obtained with the poly(A) tail sample alone are shown in the lower panel. Single-nucleotide resolution was achieved in both separation results and maintained throughout the entire poly(A) tail profile. The arrow in the upper panel indicates the 120 nt full-length size marker comigrating with one of the poly(A) tail peaks in the sample, indicating the length of this poly(A) tail peak is 120 nt. This 120 nt peak and the 121 nt peak are the most abundant poly(A) tail species in this sample, consistent with the theoretical 120 nt poly(A) tail length design by TriLink. This sample's poly(A) tail profile included a length distribution from 97 nt to 156 nt. This heterogeneity in poly(A) length may be caused by transcription slippage of the RNA polymerase used to synthesize the FLuc mRNA, as indicated previously.3
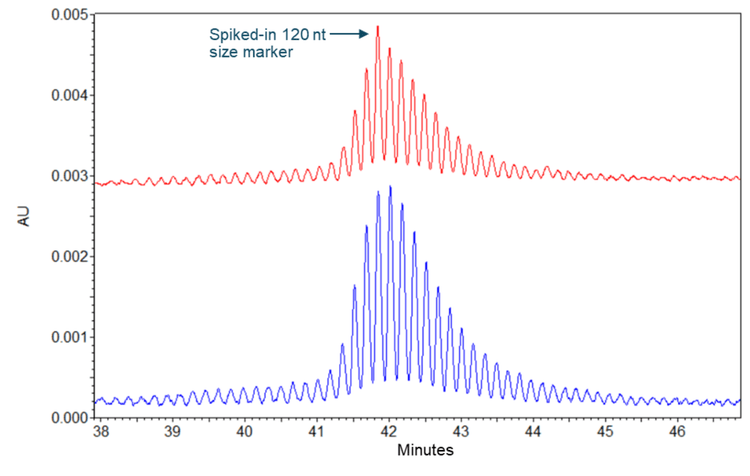
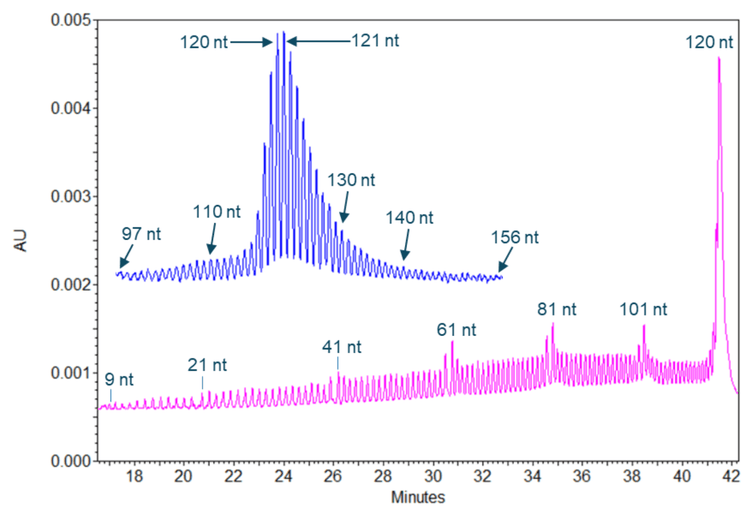
Single-nucleotide resolution maintained in the size range of 9 nt to 156 nt
Figure 5 shows results obtained with a 120 nt size marker (in the pink trace) and with poly(A) tails from the TriLink FLuc mRNA (in the blue trace). The 120 nt full-length product for this size marker and n-1 species down to the size of 9 nt were well separated with single nucleotide resolution. The single-nucleotide resolution was also achieved in analyzing the poly(A) tails from the TriLink FLuc mRNA with a length distribution of 97 to 156 nt. These results demonstrate a sustained single-nucleotide resolution in the size range of 9 nt to 156 nt. For mRNA-based therapeutics, it was shown that a poly (A) tail length of approximately 100 nt is optimal to minimize degradation.4 However, it has been reported that short poly(A) tails are associated with highly expressed, well-translated genes. 5 Some very stable natural transcripts, such as beta-actin, were shown to have a short poly(A) tail of less than 30 nucleotides. Therefore, the poly(A) tail analysis workflow by CGE-UV is suitable for analyzing mRNA-based therapeutics and natural transcripts.
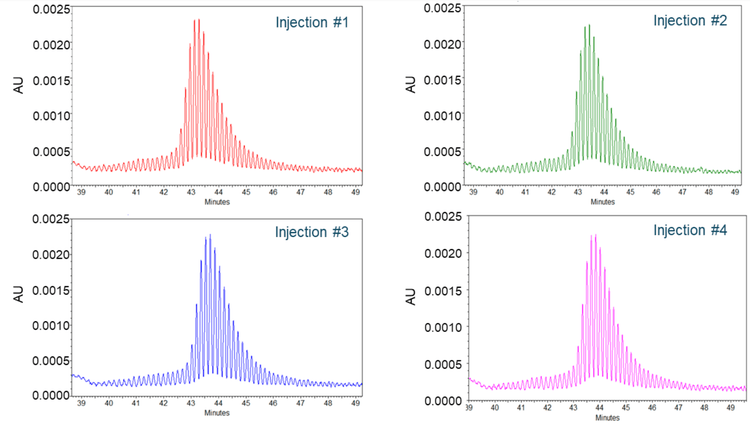
Excellent assay repeatability
To assess the assay repeatability, poly(A) tails from the TriLink FLuc mRNA sample were injected 4 times, and the results are shown in Figure 6. Each panel displays the electropherogram from 1 injection. The poly(A) tail profiles in all 4 injections were consistent, with 120 nt and 121 nt peaks as the most abundant species, and single nucleotide resolution was maintained throughout the profile for each injection. The CVs for the migration times and the corrected peak area percentage (CPA%) of the 120 and 121 nt species across 4 injections were better than 0.60% and 2.50%, respectively, demonstrating excellent assay repeatability.
Peak quantitation of poly(A) tail species
Maintaining single-nucleotide resolution throughout the entire poly(A) tail profile enabled the execution of peak integration. The CPA% of each poly(A) tail species of the TriLink FLuc mRNA sample was determined using the 32 karat software, and results were summarized in Figure 7. The poly(A) tail distribution of the TriLink FLuc mRNA covers 60 fragments in a size range of 97 nt to 156 nt. Each fragment peak had a signal-to-noise ratio of 5 or higher. The most abundant 2 species (dark green) are the 120 nt and the 121 nt fragments, each representing about 9%. These 2 species, together with 17 additional species with lengths ranging from 116 nt to 132 nt, cover about 80% of all poly(A) tails. This quantitative information adds value to the characterization and quality monitoring of mRNA-based therapeutics.
Conclusion
- Single-nucleotide resolution was achieved with the CGE-UV workflow, enabling accurate and direct measurement of poly(A) tail length
- The single-nucleotide resolution was sustained across the entire poly(A) tail profile in the size range of 9 to 156 nt, facilitating peak integration of each individual poly(A) tail species
- Excellent assay repeatability was demonstrated by multiple injections of the same poly(A) tail sample with CVs for the migration times and the CPA% of the 120 and 121 nt species across 4 injections better than 0.60% and 2.50%, respectively
- Feasibility of the CGE-UV as a QC-friendly workflow for poly(A) tail analysis was demonstrated with simple and fast sample preparation and ready-to-use ssDNA 100-R kit on a 21 CFR Part 11 compliance-ready platform
- Potential of CGE-UV workflow for development of QC assay for poly(A) tail analysis with simple and fast sample preparation and ready-to-use ssDNA 100-R kit on a 21 CFR Part 11 compliance-ready platform
References
- Di Grandi et al., (2023) A single-nucleotide resolution capillary gel electrophoresis workflow for poly(A) tail characterization in the development of mRNA therapeutics and vaccines. J Pharm Biomed Anal. Nov 30; 236:115692. PMID: 37696189.
https://doi.org/10.1016/j.jpba.2023.115692 - ssDNA 100-R kit. SCIEX application guide, RUO-IDV-05-11137- A.
- Molodtsov et al., (2014) The presence of an RNA:DNA hybrid that is prone to slippage promotes termination by T7 RNA polymerase. J. Mol. Biol. 426:3095–3107. PMID: PMID: 24976131
https://doi.org/10.1016/j.jmb.2014.06.012 - Schlake et al., (2012) Developing mRNA vaccine technologies. RNA Biol. 9:1319–30. PMID: 23064118
https://doi.org/10.4161/rna.22269 - Lima et al., (2017) Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol 24:1057– 1063. PMID: 29106412
https://doi.org/10.1038/nsmb.3499


