Introduction
This technical note presents a straightforward workflow to characterize and relatively quantify the 5’-capping structures of therapeutic messenger RNA (mRNA). It enables scientists to assess different 5’-capping structures with high confidence based on high-quality accurate mass data.
Although mRNA has been studied with regard to its medical use for several decades, the SARS-CoV-2 pandemic has led to an unprecedented acceleration in the maturity of the platform. Products based on mRNA have broad potential for use as therapeutics, in applications such as cancer treatments and vaccines. Since mRNA leverages the translational machinery of the host cells, there is a significantly reduced risk of unwanted post-translational modifications that can plague protein therapeutics. Nonetheless, mRNA-based products have other critical quality attributes (CQA) that directly impact both efficacy and safety. Key among these is the 5'-capping of the mRNA.1 Both the presence and specific type of 5'-cap of in vitro transcribed (IVT) mRNA have a direct impact on translational efficiency.2 Traditional biophysical techniques such as denatured gel electrophoresis or total hydrolysis have very limited utility for assessing 5' capping due to factors such as insufficient resolution, radioactive contamination and poor sensitivity. Functional assays, such as protein expression in cellular assays, are very time-consuming and can be highly confounded by different cap chemistries.3 As the development needs for mRNA products increase, the need for an easy-to-use, high-throughput, highly reproducible method for assessing mRNA 5'-capping increases simultaneously.4-6 Previously, a LC-MS method has been reported to analyze pre-defined 5’ fragments of synthetic mRNA.7
Herein, an LC-MS method for the reliable identification and relative quantitation of different mRNA capping structures was developed. The method can distinguish between uncapped and multiple different capped 5’-ends of mRNA simultaneously, addressing the need for characterizing this CQA.
Key features of mRNA capping assessment
- Fast and reliable identification of the 5’-end structure of IVT mRNA species
- Straightforward and fully customizable quantitation of mRNA capping efficiency through reconstructed masses in SCIEX OS software
- High confidence in the identification of main and low-abundance species based on high-quality accurate mass data in negative polarity
Methods
Sample preparation: An undisclosed, 5’-capped mRNA and its uncapped version produced via IVT were used after digestion. In order to determine the linear response of the assay, 0, 10 and 50% (mol:mol) of digested, uncapped mRNA samples were spiked into digested, capped mRNA samples.
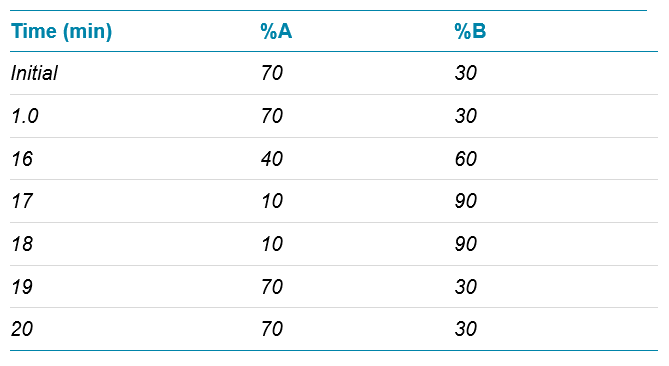
Chromatography: An ACQUITY UPLC H-class PLUS system (Waters) equipped with an ACQUITY UPLC Oligonucleotide BEH C18 column (2.1 mm×50 mm, 1.7 µm, 130Å) was used. Mobile phase A was 15mM diisopropylethylamine (DIEA) with 100mM 1,1,1,3,3,3-hexafluoro isopropanol (HFIP) in water. Mobile phase B was 15 mM DIEA with 100 mM HFIP in 50:50 (v/v) methanol/water. The gradient used is shown in Table 1. The column temperature was held at 60°C and a flow rate of 300 µL/min was used. Unless described otherwise, an injection volume of 5 µL was employed, resulting in approximately 10 pmol (70 ng) of digested mRNA product on column.
Overview
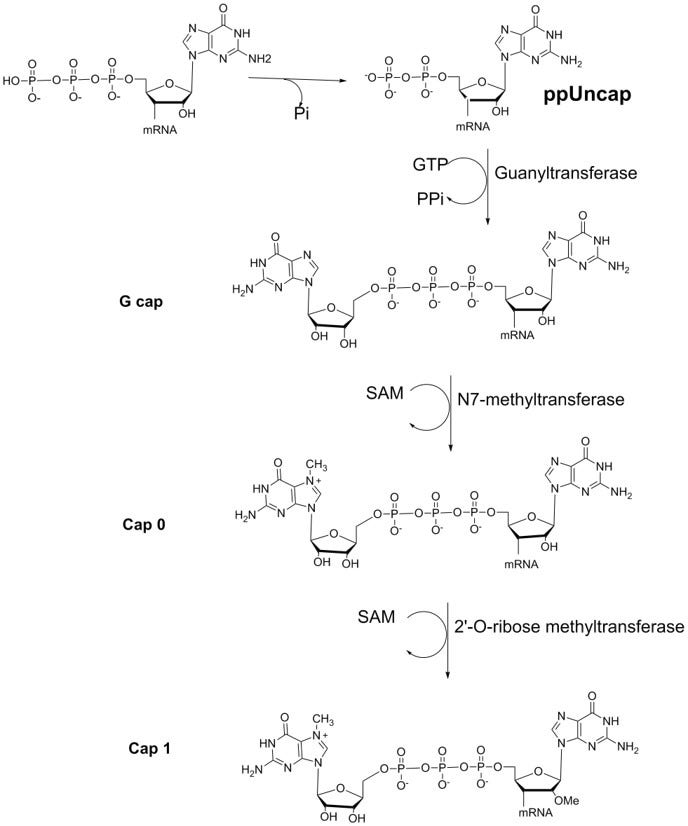
5’-capping of IVT mRNA is necessary to maintain the biological function of the mRNA. Therefore, it is considered a critical quality attribute (CQA).3 A typical enzymatic mRNA capping process involves three enzymatic reactions (Figure 2). The capping process results in a single nucleotide addition at the 5’ end (Cap1 in Figure 2). Since a typical therapeutic mRNA consists of thousands of nucleotides, traditional denatured gel electrophoresis does not provide enough resolution to separate the uncapped from the capped mRNA for quality control purposes. Full-length mRNA also surpasses the size range of typical LC-MS-based workflows for oligonucleotide characterization. Other reported methods such as cell-based functional assays or radiolabeling either suffer from low throughput and high variability or require the modification of the product, which is not suitable for the development of therapeutic mRNA. To address these challenges, an LC-MS workflow suitable for the detection of digested mRNA was developed. With this workflow, a detailed characterization of the 5’-capping can be performed and the efficiency of the capping can be quantified simultaneously.
Identification of capping structures
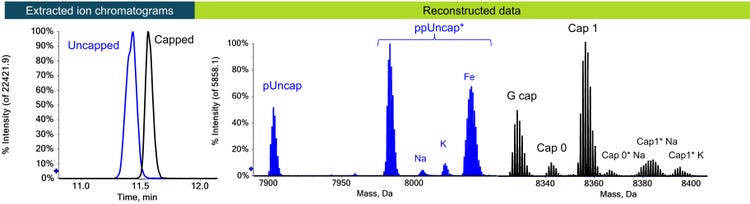
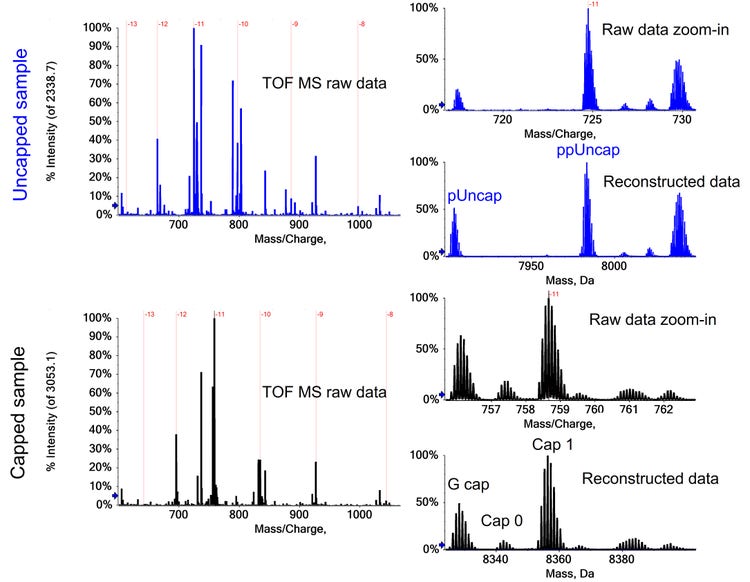
The LC gradient was optimized to allow for the separation of the uncapped and capped species (Figure 1A). Different amounts of phosphorylation as well as metal adducts were identified for the capped and uncapped mRNA based on the reconstructed accurate mass data (Figure 1B and C). The separation (Figure 1A) confirmed that the uncapped species were not a product of in-source fragmentation, but present in the sample.
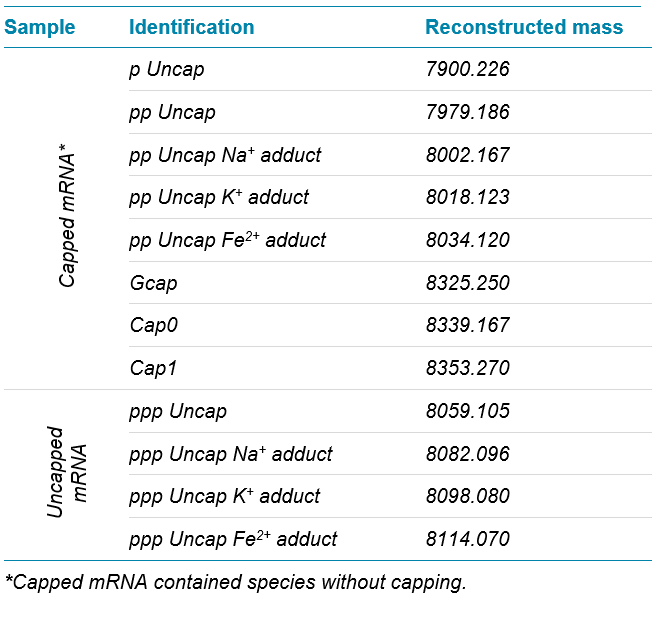
As a control, an uncapped sample was digested and analyzed in parallel. An uncapped triphosphate-containing product was observed with low levels of monophosphate or diphosphate versions as expected. Table 3 provides a summary of all identified species. All species showed excellent resolution for raw and reconstructed data, and the comparability of reconstructed to raw data was confirmed (Figure 3).
Relative quantification of capping efficiency
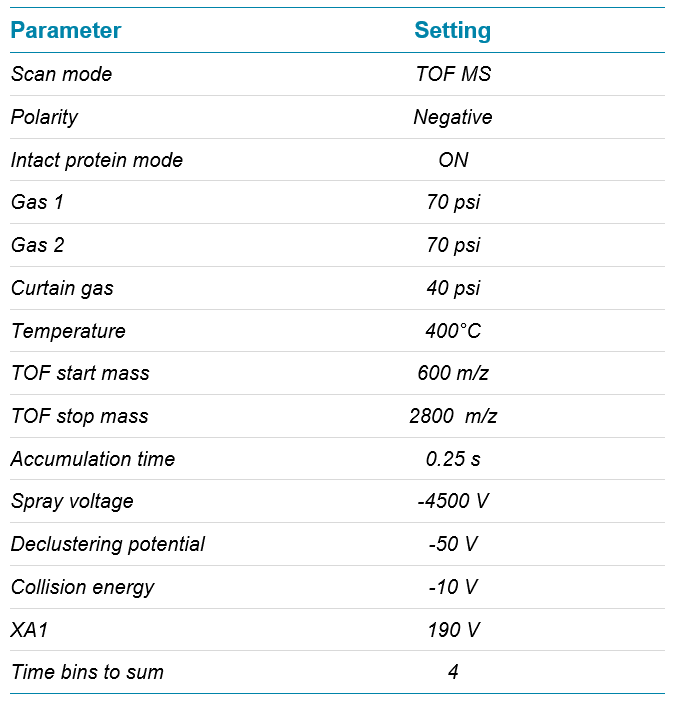
To assess the capping efficiency, the intact quantification workflow in SCIEX OS software was used, as described earlier.9-10 In brief, raw TOF MS spectra were reconstructed and mass peaks were integrated using the Analytics module in SCIEX OS software. A customized formula was used to calculate peak area ratios of each species against the sum of all species. All identified adducts were taken into account for highest accuracy. With that approach the capping efficiency was determined to be 43.2%. The quantification can be customized to allow assessment of quantities for each species individually.
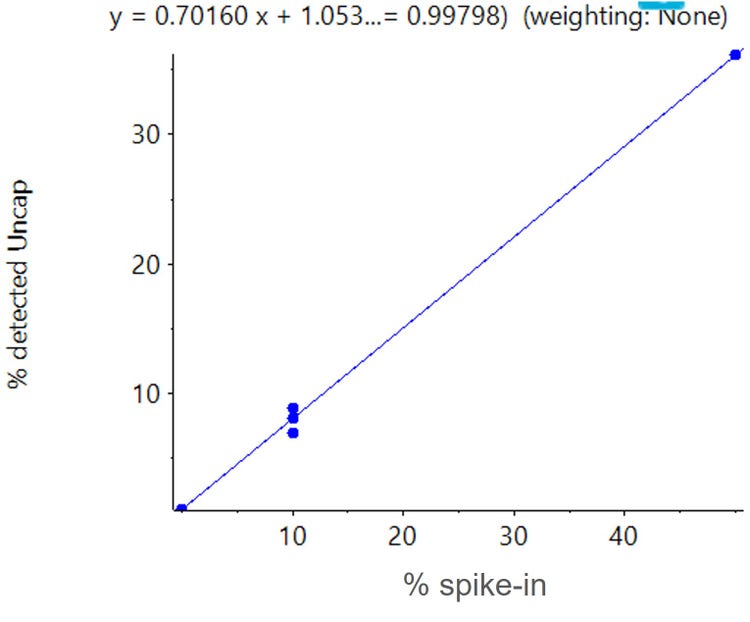
As a separate experiment, the reliability of the quantification was examined. Different amounts of the uncapped mRNA sample (0, 10 and 50%) were spiked into the capped sample prior to digestion. The digestion products were analyzed using the same LC-MS workflow (Table 1 and 2). The species contributing to the uncapped sample, specifically the triphosphate-containing version of the cleaved product, were reconstructed, integrated and their detected ratios were plotted against the known, spiked-in ratios using SCIEX OS software. A linear response was observed between the two values (Figure 4), confirming the quantitative accuracy of the approach.
Conclusions
- Separation of capped and uncapped species of IVT mRNA was achieved using low ion-pairing concentrations
- Confident identification of the capped mRNA and multiple process-related byproducts in the sample mixture was enabled by high-quality accurate mass TOF MS data
- Highly efficient and reliable data reduction and easy determination of capping efficiency were provided by the intact quantitation workflow in SCIEX OS software
- Confident determination of capping efficiencies was confirmed by assessment of the linearity of the LC-MS response
References
- Ramanathan A, Robb GB, Chan SH (2016). mRNA capping: biological functions and applications. Nucleic Acids Res. 44(16): 7511-26.
- Pardi N, Hogan MJ, Porter FW, Weissman D (2018). mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov.17(4): 261-279.
- Grudzien-Nogalska E, Kowalska J, Su W et al. (2013). Synthetic mRNAs with superior translation and stability properties. Methods Mol Biol.969:55-72.
- Kwon H et al. (2018). Emergence of synthetic mRNA: In vitro synthesis of mRNA and its applications in regenerative medicine, Biomaterials 156:172.
- Sahin U, et al (2014). mRNA-based therapeutics--developing a new class of drugs. Nat Rev Drug Discov. 13, 759.
- Riley JA.CMC considerations for mRNA based therapeutics: Analytical approaches to characterize critical quality attributes. Intertek.
- Beverly, M.; Dell, A.; Parmar, P.; Houghton, L. Label-Free Analysis of mRNA Capping Efficiency Using RNase H Probes and LC-MS. Anal. Bioanal. Chem. 2016, 408, 5021–30.
- Selina T. M. Monn et al (2005) Investigation of Metal-Oligonucleotide Complexes by Nanoelectrospray Tandem Mass Spectrometry in the Positive Mode J Am Soc Mass Spectrom, 16:370–378.
- Robert L. Hettich (1999). Formation and Characterization of Iron–Oligonucleotide Complexes with Matrix-Assisted Laser Desorption/Ionization Fourier Transform Ion Cyclotron Resonance Mass Spectrometry, J Am Soc Mass Spectrom, 10: 941.
- Charge variant liability analysis using the SCIEX flexible solution for MAM, SCIEX technical note, RUO-MKT-02-11828-A.