Abstract
This technical note demonstrates the power of electron activated dissociation (EAD) to confidently identify and characterize advanced glycation end products (AGEs) in protein therapeutics. EAD resulted in excellent fragmentation and complete coverage of AGEs, leading to high confidence in sequence identification and localization of AGE moieties.
Introduction
Protein therapeutics with glycation can undergo further degradation to produce AGEs in vivo. 1-2 AGEs might cause discoloration of therapeutic products and induce adverse immune responses. 1-4 Despite their significance to product quality, AGEs are poorly characterized because they have low abundances and are difficult to fragment using traditional collision-based MS/MS approaches. 2-4 In comparison, EAD can provide excellent fragmentation of glycated peptides, 4 allowing the accurate localization of the glycation moiety to differentiate between positional isomers.
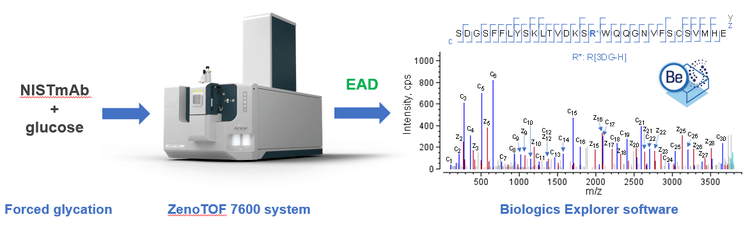
In this technical note, EAD was employed to characterize the AGEs produced from forced glycation of NISTmAb (Figure 1). EAD enabled the precise localization of the AGE moieties, even in the presence of glycation and/or glycosylation. To our knowledge, this is the first report on the characterization of AGEs using an alternative fragmentation approach.
Key features of EAD for the comprehensive characterization of glycation and AGEs
- Confident identification: EAD enables excellent fragmentation of glycated peptides and AGEs, leading to high confidence in sequence identification
- Accurate localization: The glycation or AGE moieties are retained in EAD, allowing unambiguous localization of these labile modifications and differentiation between isomeric Lysand Arg-associated AGEs
- High quality: The Zeno trap provides a 5–10-fold increase in the detection of MS/MS fragments, improving EAD data quality for glycated peptides or AGEs at varied concentrations
- Fast and flexible: EAD can be operated in data-dependent acquisition (DDA) or MRMHR mode with a fast scanning rate (~20 Hz in DDA mode) and with the ability to tune electron kinetic energy (KE)
Methods
Sample preparation: Three vials of 10 µg/µL NISTmAb (RM 8671, NIST) were mixed with 500mM glucose (Sigma-Aldrich) and incubated at 60ºC for 4 days. The solution turned yellowbrown following forced glycation. The stressed samples were denatured in guanidine hydrochloride, reduced with dithiothreitol and alkylated with iodoacetamide. Buffer exchange was then performed using Bio-Spin 6 columns (Bio-Rad Laboratories). Three vials of sample were separately digested (at 37ºC for 3 hours) using 3 different enzymes, including trypsin/Lys-C mix (Promega), chymotrypsin (Promega) and Glu-C (Promega). The final digests were injected in 10 µL aliquots (~10 µg) for EAD analysis.
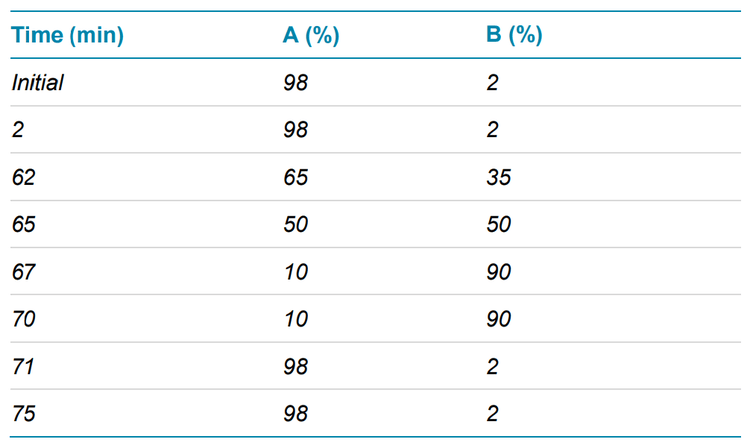
Chromatography: The peptides were separated with the gradient displayed in Table 1 using an ACQUITY CSH C18 column (2.1 × 150 mm, 1.7 µm, 130 Å, Waters). A flow rate of 0.25 mL/min was used for the peptide separation. The column was kept at 60ºC in the column oven of an ExionLC system (SCIEX). Mobile phase A was 0.1% formic acid (FA) in water and mobile phase B was 0.1% FA in acetonitrile.
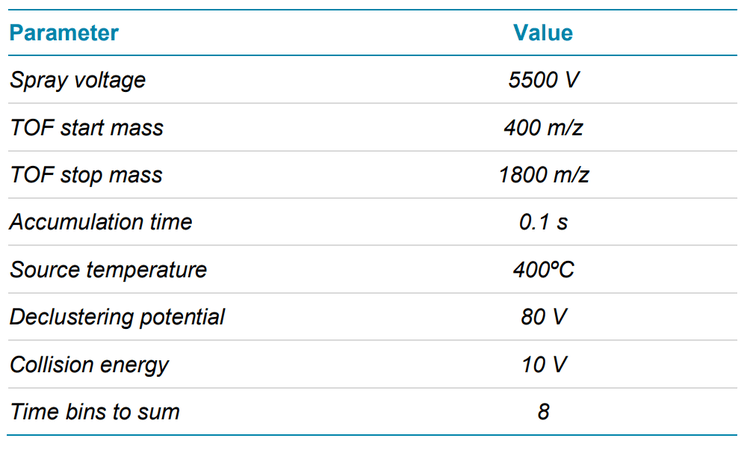
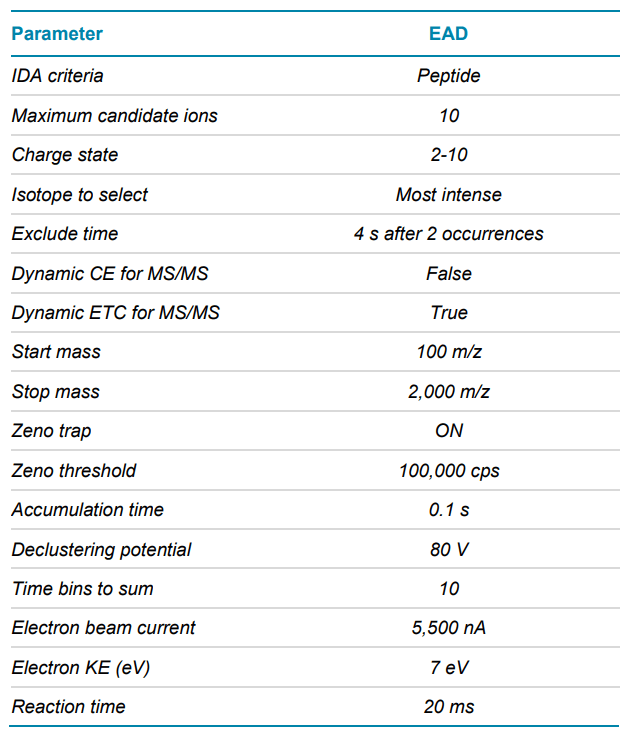
Mass spectrometry: EAD DDA data were acquired in SCIEX OS software using the ZenoTOF 7600 system. The key TOF MS and MS/MS settings are listed in Tables 2 and 3, respectively.
Data processing: EAD data were analyzed using the peptide mapping workflow templates within the Biologics Explorer software. A maximum of 4 hexose modifications at Lys or Arg residues per peptide sequence was allowed in peptide mapping. The AGE modifications on Lys residues that were considered for data analysis included Hex-1H2O (+144.04 Da), Hex-2H2O (+126.03 Da), carboxymethyl (CML, +58.01 Da), carboxyethyl (CEL, + 72.02 Da), furosine or pyrraline (FRS/PRL, +108.02 Da). The AGE modifications on Arg residues that were considered for data analysis included 3DG-H (+144.04 Da), dihydropyrimidine (DHPM, +126.03 Da), glyoxal hydroxyimidazolone (G-OH, +37.98 Da) and glycoal hydroimidazolone (G-H, +39.99 Da). Oxidations at Met, His and Trp residues were set as variable modifications.
Glycation and AGEs of mAbs
Glycation of a recombinant monoclonal antibody (mAb) occurs when the proteins are exposed to reducing sugars, such as glucose, during fermentation or storage.1-2 Glycated mAbs can undergo further degradation to form AGEs in vivo or under stress conditions. 1-2 While glycation does not change the color of mAbs, AGEs are linked to the discoloration of the product.3-4 Additionally, AGEs might trigger the expression of AGE-specific receptors and induce adverse immune responses.1-2 Given these safety concerns, glycation and AGEs must be fully characterized and closely moonitored during the lifecycle of a protein therapeutic.
Detailed characterization of AGEs by LC-MS has been scarce3 due to the low abundances of glycation and AGEs and the poor fragmentation of these species with traditional collision-based MS/MS approaches. In this work, NISTmAb was thermally stressed in the presence of glucose for 4 days to promote the formation of AGEs, which were then characterized using EAD. The results described in this technical note enhance our understanding of AGEs and highlight the power of EAD to comprehensively characterize these challenging modifications.
Comprehensive characterization of AGEs from NISTmAb with forced glycation
As described in a previous technical note, 3 enzymes with complementary cleavage patterns, including trypsin/Lys-C, chymotrypsin and Glu-C, were employed to facilitate the complete characterization of glycated peptides that resulted from the forced glycation of NISTmAb. 5 Here, digestion of glycated NISTmAb using these enzymes produced different length peptides that carried the same AGE modification on the same site (Lys or Arg). High-quality EAD data of these peptides, regardless of the length, provided additional confidence in identifying the AGEs.
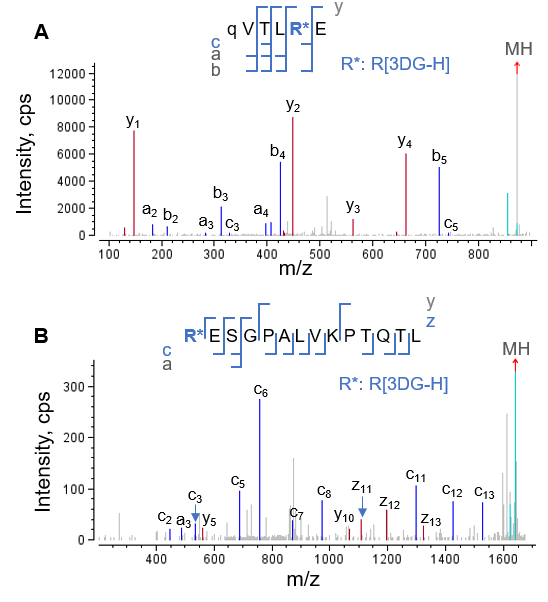
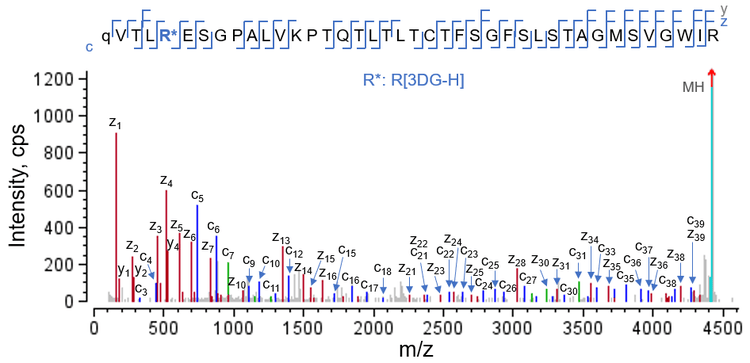
EAD MS/MS spectra of 3 N-terminal HC peptides that contained the same AGE modification (3DG-H at Arg5 ) are displayed in Figures 2 and 3. Glu-C digestion of glycated NISTmAb produced an N-terminal peptide from HC[1-6], with 6 amino acid residues (Figure 2A). In contrast, the chymotrypsin and trypsin/Lys-C digestions produced the longer peptides HC[5-18] and HC[1-40], respectively. Despite a drastic difference in peptide length, EAD resulted in complete or nearly complete fragmentation of these peptides with AGE modifications, enabling confident peptide identification and accurate localization of the 3DG-H moiety to Arg5 (Figures 2 and 3).
One of the challenges faced when characterizing glycation or AGE modifications is the differentiation between positional isomers that carry modifications with identical mass values on different amino acid residues. For example, the differentiation between isomeric AGEs containing a Hex-1H2O modification on a Lys residue or a 3DG-H on an Arg residue (+C6H8O4 or +144.04Da for both AGE modifications), is challenging. EAD can address this challenge because it yields excellent fragmentation of peptides with AGEs while retaining the labile modifications. The accurate localization of 3DG-H to Arg5 in the AGEs described here (Figures 2 and 3) eliminated the possibility of a positional isomer in which a Lys residue was modified with Hex1H2O.
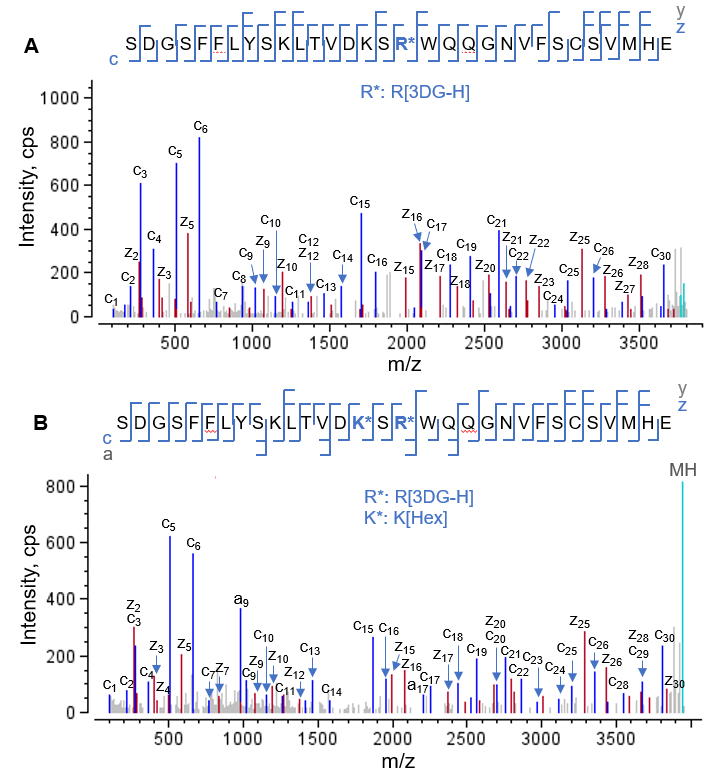
Excellent fragmentation of AGEs using EAD was also achieved for those containing multiple modifications. Figure 4 displays EAD MS/MS spectra of the peptide HC[403-433] modified with a 3DG-H moiety (Figure 4A) and the same peptide carrying both a 3DG-H and a glycation moiety (Figure 4B). The detection of a nearly complete series of c/z fragments for 2 AGEs allowed accurate localization of 3DG-H individually (Figure 4A) or simultaneously with the glycation moiety (Figure 4B).
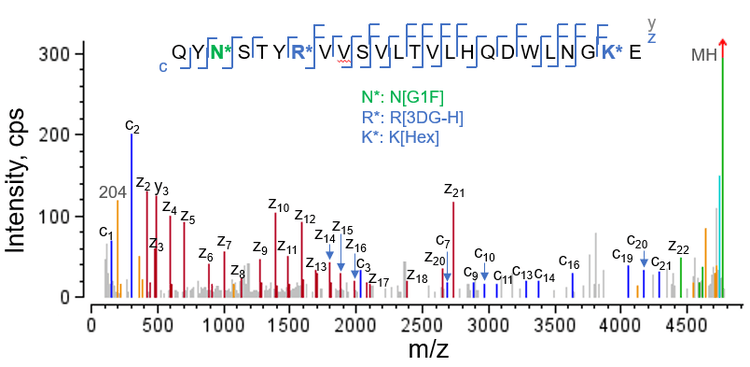
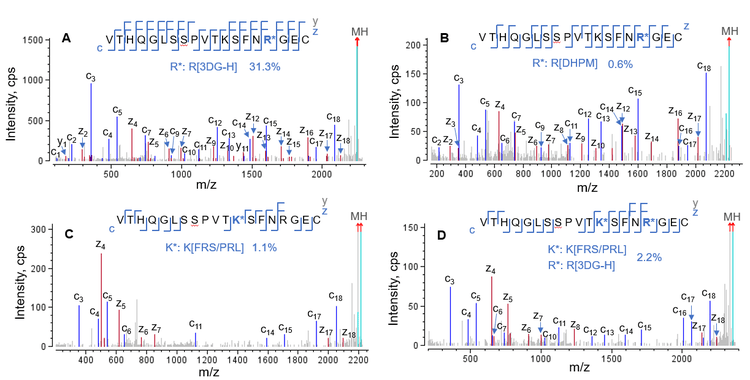
Figure 5 displays the EAD spectrum of an HC peptide carrying 3 different labile modifications, including 1 glycation residue (Hex), 1 AGE moiety (3DG-H) and 1 glycan (G1F). Despite the complex modification pattern of this peptide, EAD led to nearly complete coverage of the sequence containing the 3 labile moieties, allowing precise localization of each modification. For example, the detection of 2 complementary pairs of c2/z22 and c3/z21 fragments confirmed the G1F glycosylation on Asn3 and the detection of c7/z18 ions localized 3DG-H to Arg7 .
Excellent EAD data enabled the confident identification of the same sequence modified with different AGE moieties. Figure 6 shows the EAD spectra of LC[195-213] modified with different AGE moieties, including 3DG-H (Figure 6A), DHPM (Figure 6B), FRS/PRL (Figure 6C) and the combination of 3DG-H and FRS/PRL (Figure 6D). These 4 AGEs were present at different levels, ranging from a relative abundance of >30% for 3DG-H (Figure 6A) to as low as 0.6% for DHPM (Figure 6B). Regardless of the type and relative abundance of these AGEs, EAD produced excellent MS/MS fragmentation to confidently identify and localize the AGE moieties.
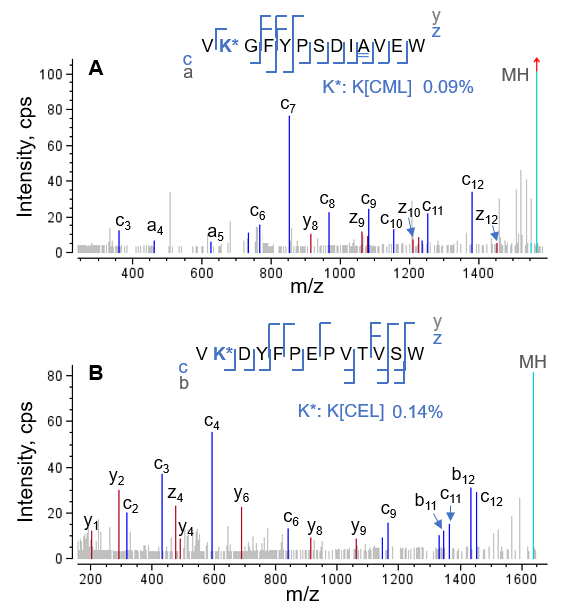
Additionally, EAD DDA is a highly sensitive approach because the use of the Zeno trap provides a 5‒10-fold increase in the detection of MS/MS fragments. Two examples that highlight the ability of EAD to characterize AGEs at low levels are displayed in Figure 7. The CML on peptide HC[372-384] (Figure 7A) and CEL on peptide LC[149-161] (Figure 7B) were present at ~0.1% relative abundance, whereas the corresponding glycated species were present at 85-95% relative abundance (data not shown). Despite the low abundances of CML and CEL, EAD DDA produced enough sequence ions to confidently identify the 2 species and localize the AGE moieties.
Summary of AGEs identified by EAD
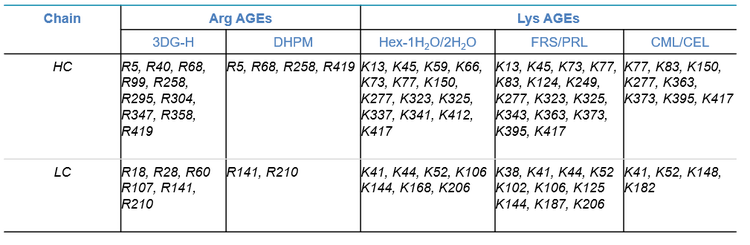
Forced glycation of NISTmAb resulted in the formation of numerous glycated species and AGEs. The previous technical note described selected examples of glycated peptides identified by EAD. 5 Table 4 summarizes the major AGEs and their modification sites identified from glycated NISTmAb using EAD in this work. The dominant AGE modification identified in the forced glycation samples was 3DG-H on the Arg residues (Table 4). Other common AGE modifications observed included Hex1H2O/2H2O and FRS/PRL at Lys and DHPM at Arg. The AGEs with CML or CEL were only present at low levels.
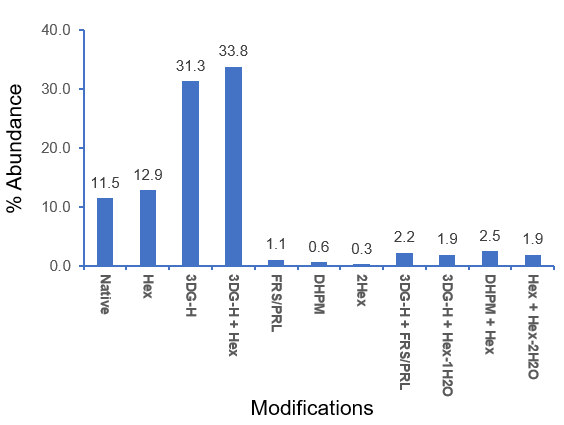
Figure 8 shows the relative abundances of the glycated species and AGEs identified for the peptide LC[195-213] from the chymotrypsin digestion. Forced glycation of NISTmAb resulted in extensive modification of this LC peptide, with 88.5% of the peptide modified with 1 or 2 glycations and/or AGE moieties. The dominant AGEs identified contained 1 3DG-H moiety with or without 1 glycation residue (Hex). The relative abundances of these 2 AGEs (31-34%) were much higher than those of other species, including the glycated peak (12.9%), indicating the prevalence of the 3DG-H modification. The relative quantification of glycation and AGE modifications will be described in another technical note.
In summary, EAD is powerful for the comprehensive characterization of glycation and AGEs, which are challenging modifications to characterize using traditional collision-based MS/MS approaches. EAD resulted in excellent fragmentation of AGEs to confidently identify peptide sequences. Additionally, this method preserved AGE moieties to accurately localize these labile modifications and enable differentiation between positional isomers.
Conclusion
- EAD is a powerful tool for in-depth characterization of glycation and AGEs, which are important for the quality of protein therapeutics but challenging to characterize using traditional collision-based MS/MS approaches
- EAD led to nearly complete fragmentation of AGEs while retaining the AGE moieties for confident sequence identification, accurate localization of labile modifications and differentiation between positional isomers
- EAD enabled the confident characterization of AGEs of different lengths and present at abundances as low as ~0.1%
- The major AGE modifications identified from forced glycation samples of NISTmAb included Arg-derived 3DG-H and DHPM and Lys-derived Hex-1H2O/2H2O, FRS/PRL and CML/CEL, among which 3DG-H was the dominant moiety.
References
- Bingchuan Wei et al. (2017) Glycation of antibodies: Modification, methods and potential effects on biological functions. MABS. 9(4): 586-594.
- Anna Robotham and John Kelly (2020) LC-MS characterization of antibody-based therapeutics: recent highlights and future prospects. Approaches to the Purification, Analysis and Characterization of AntibodyBased Therapeutics. Chapter 1: 1-33.
- Margaret Butko et al. (2014) Recombinant antibody color resulting from advanced glycation end product modifications. Anal. Chem. 86(19): 9816-9823.
- Alexandre Ambrogelly (2021) The Different Colors of mAbs in Solution. Antibodies. 10(2): 21.
- Comprehensive characterization of glycation in protein therapeutics using electron activated dissociation (EAD). SCIEX technical note, RUO-MKT-02-15020-A.