Comprehensive mapping of disulfide linkages of adalimumab using electron activated dissociation (EAD) and collision induced dissociation (CID)
Mona Hamada1, Zoe Zhang2 and Amandine Boudreau1
1SCIEX, Canada; 2SCIEX, USA
Abstract
This technical note describes a comprehensive mapping of expected and scrambled disulfide linkages and free thiols in adalimumab using EAD and CID. Data were acquired on the ZenoTOF 7600 system with SCIEX OS software, and automated data interpretation was performed using Biologics Explorer software.
Disulfide linkages are recognized by regulatory agencies such as the FDA, EMA and ICH as critical quality attributes (CQAs) of protein biotherapeutics.1,2 Reduction and scrambling of the disulfide linkages can occur during manufacturing and storage, disrupting the proper folding of biotherapeutics and consequently impacting their functions.1,2 Therefore, accurate mapping of disulfide linkages and quantitative assessment of their variants are crucial to ensure the safety, efficacy, and stability of the drug product.
CID is the most widely adopted fragmentation mode in LC-MS bottom-up peptide mapping analysis.3 However, CID may not fragment disulfide linkages, and their presence can further hinder effective peptide backbone fragmentation, resulting in limited sequence information.2,3 In contrast, EAD effectively cleaves the disulfide linkages, further empowering its efficient peptide backbone fragmentation.2,3 EAD is an exceptionally powerful tool that allows the comprehensive mapping of the cysteine residues and the peptides involved in these complex linkages.3
The data presented in this technical note shows the in-depth characterization of the complex disulfide linkage structure of adalimumab, including the identification of impurities such as free thiols and scrambled disulfide peptides.
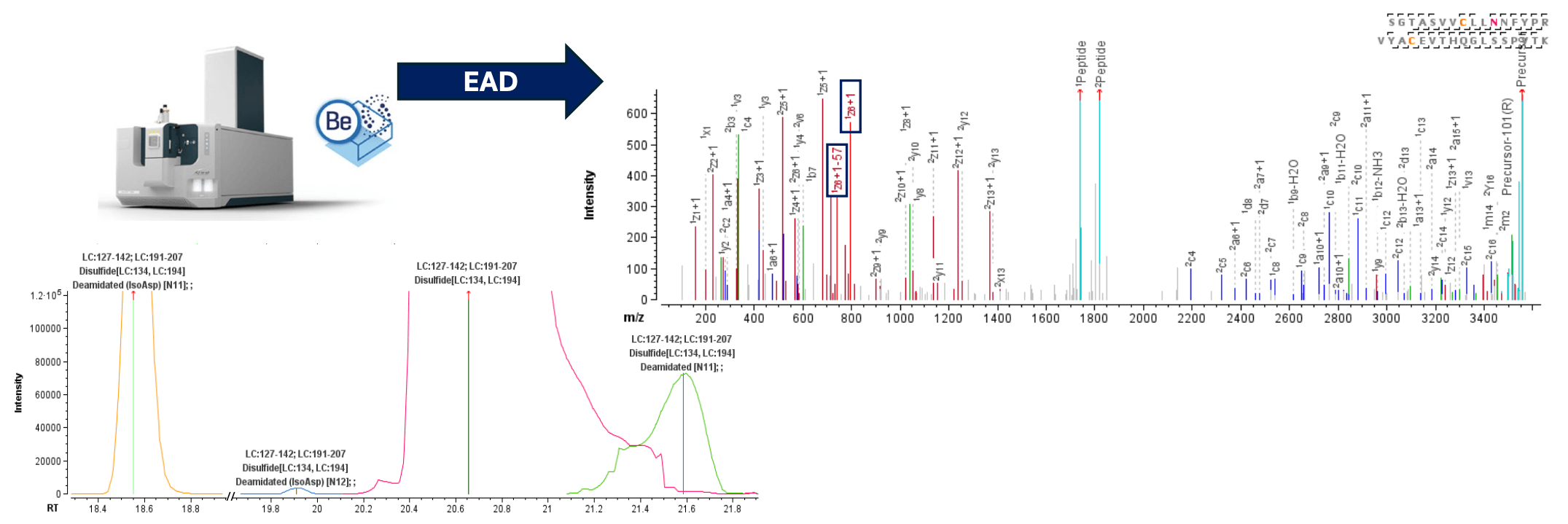
Figure 1. Characterization of disulfide linkages and PTMs on the same peptideusing EAD with the ZenoTOF 7600 system. An intrachain disulfide-linkedpeptide from the light chain was chromatographically resolved from its deamidated forms (left panel). The highly sensitive EAD method offered by the ZenoTOF 7600 system led to rich fragmentation spectra of the disulfide-linked peptides (right panel). The detection of diagnostic fragments produced by EAD confirms the isoAsp formation at positions N11 (Asn137) and N12 (Asn138) following deamidation.
Key features of disulfide linkage analysis using Biologics Explorer software
- Confident identification: High-quality MS/MS data obtained by EAD and CID fragmentation allows solid sequence confirmation and characterization of expected and scrambled disulfide linkages and free thiols.
- Accurate localization: Post-translational modifications (PTM), including amino acid isomerization, can be unambiguously localized on disulfide-linked peptides in the same run.
- Powerful, streamlined solution: EAD combined with superior data processing algorithms in Biologics Explorer software provides a complete solution for challenging disulfide linkages analysis.
Introduction
Adalimumab is a recombinant human IgG1 monoclonal antibody that prevents the interaction of tumor necrosis factor (TNF) with TNF receptors. IgG1 antibodies are stabilized by 16 disulfide linkages (12 intrachain and 4 interchain) formed between all 32 Cys residues.1,4 Both intra- and inter-chain disulfide bonds are critical for establishing the tertiary and quaternary structure of biologics.4 Normally, Cys residues pair with their correct partner residues. However, under stress conditions such as heat, radicals, improper pH or agitation, unexpected linkages can be formed via free thiols, resulting in potential detrimental effects on the physicochemical and biological properties of biologics.1
Methods
Sample preparation: Adalimumab was denaturated with 7.2M guanidine hydrochloride in 50mM Tris buffer (pH 7.0). Free cysteine was capped with 5mM iodoacetamide. Protein digestion was performed with trypsin/Lys-C enzyme at 30°C for 16 hours at pH 7. The reaction was stopped with 1% formic acid.
Chromatography: 4 μg (CID) or 8 μg (EAD) of the trypsin/Lys - C digest were separated with a CSH C18 column (2.1 × 100 mm, 1.7 μm, 130 AÅ, Waters) using an ExionLC AD system. The mobile phase A consisted of water with 0.1% formic acid, while the mobile phase B was 0.1% formic acid in acetonitrile.
A gradient profile was used at a flow rate of 300 μL/min (Table 1). The column temperature was maintained at 50°C.
Table 1. LC gradient for peptide separation.
Mass spectrometry: Data were acquired on the ZenoTOF 7600 system in positive ionization mode with a DDA method using SCIEX OS software. EAD and CID MS/MS data were acquired in separate injections. Detailed method parameters are described in Table 2.
Table 2. DDA parameters using CID and EAD.
Data processing: EAD and CID DDA data were analyzed in Biologics Explorer software version 5.0 using the Pepmap_Extended_Be5.0 workflow. The following fixed disulfide linkages were specified in the first peptide mapping activity node: [HC:22, HC:96], [HC:148, HC:204], [HC:224, LC:214], [HC:230, HC:230], [HC:233, HC:233], [HC:265, HC:325], [HC:371, HC:429], [LC:23, LC:88] and [LC:134, LC:194]. The second peptide mapping activity node was set for the de-novo mapping of unexpected disulfide linkages. For all other parameters, the default workflow settings were used.
Figure 2. EAD (top) and CID (bottom) MS/MS spectra of the disulfide-linked peptide SLRLSCAASGFTFDDYAMHWVR=AEDTAVYYCAK, Cys22=Cys96, an intrachain disulfide bond from the heavy chain.
Identification of expected disulfide linkages
The ZenoTOF 7600 system introduces an alternative fragmentation mechanism, EAD. In addition, the Zeno trap improves the MS/MS sensitivity up to 7-fold and further enhances the detection of fragment ions generated by EAD or CID. Consequently, rich MS/MS spectra are generated for the analysis of disulfide-linked peptides, augmenting data confidence. Combined with automated data processing using Biologics Explorer software, a streamlined solution is provided for complex disulfide linkages analysis.
The characterization results obtained from EAD and CID were compared. Both techniques successfully identified all expected disulfide linkages in adalimumab with a mass error below 8 ppm. Overall, EAD demonstrated efficient disulfide linkage cleavage during the fragmentation, leading to a higher coverage of the peptide backbone in the MS/MS spectra, especially for the longer peptides.
Figure 2 shows an example of EAD and CID MS/MS spectra for peptide SLRLSCAASGFTFDDYAMHWVR=AEDTAVYYCAK (z=+5, 753.55 m/z). The peptide, with a molecular weight of 3,762 Da, contains a disulfide bond as shown. In EAD analysis, the disulfide-linked peptide was identified in 3 different charge states (z=+4, +5, +6) with a high score of 328. Notably, EAD generates dominant fragments resulting from the cleavage of the disulfide bond, leading to efficient fragmentation of the peptide backbones. Consequently, it results in a higher amino acid sequence coverage (73%, 84% and 81% for the 3 charge states of the linked peptide, respectively). On the other hand, using CID, the disulfidelinked peptide was detected in charge state z=+5 with a lower score of 270. Due to the inability of CID to fragment the disulfide linkage, a lower amino acid sequence coverage (60%) was observed for the linked peptide.
Figure 3. EAD MS/MS spectra of the disulfide linked peptide SGTASVVCLLNNFYPR=VYACEVTHQGLSSPVTK, Cys134-Cys194 of the light chain. Asp and isoAsp can be localized and differentiated between Asn137 (top) or Asn138 (bottom) due to the signature fragment ion pair generation by EAD.
PTM characterization on disulfide-linked peptides
EAD significantly enhances the characterization and localization of post-translational modifications (PTMs) on disulfide-linked peptides, enabling comprehensive characterization of challenging PTMs such as isomerization. Figure 1 shows the chromatographic separation of SGTASVVCLLNNFYPR=VYACEVTHQGLSSPVTK, an intrachain disulfide-linked peptide formed via Cys134 and Cys194 of the light chain, along with its deamidated forms. Both Asn137 and Asn138 are possible locations for deamidation. Further, isomerization of Asp to isoAsp can happen after the deamidation. The localization and differentiation of isomers can be easily confirmed owing to the effective backbone fragmentation by EAD and its ability to generate signature fragment ions from isomers.
As shown in Figure 3, the presence of the signature fragment ion pair z• 6 and z• 6-57 confirms the deamidation at Asn 137 is a formation of isoAsp, with a mass error of less than 2 ppm. On the other hand, despite the very low abundance of the peptide observed at 19.9 min (Figure 1), EAD successfully generated the signature fragment ion pair z• 5 and z• 5-57 (with a mass error <7 ppm), further confirming the formation of isoAsp at Asn138 (Figure 3)
Identification of complex disulfide linkages
The most complex disulfide linkage of adalimumab lies in the hinge region, where the heavy chain is stabilized by 2 interchain disulfide bonds at Cys230-Cys230 and Cys233-Cys233. As such, non-reduced digest is anticipated to yield peptides connected by double or triple disulfide bonds.
Figure 4 displays the EAD and CID MS/MS spectra of 3 peptides linked by 3 disulfide bonds, producing a large peptide complex (z=+8, m/z 838.163, mass error=1.72 ppm). Notable differences were observed between the 2 fragmentation techniques for this challenging peptide. EAD exhibited a preference for cleaving the disulfide linkages, generating fragments corresponding to the individual peptides. However, these peptides were absent in the CID spectrum due to its incapacity to fragment the disulfide linkages effectively. In addition, the disulfide linkage between peptide1 (Cys224, HC) and peptide3 (Cys214, LC) was confirmed in the EAD spectrum by 2 fragment ions (1c2 and 1c3, with mass errors of -1.9 and -9.4 ppm, respectively).
Figure 4. TOF MS (left) and MS/MS (right) data for the precursor SCDKTHTCPPCPAPELLGGPSVFLFPPKPK=THTCPPCPAPELLGGPSVFLFPPKPK= SFNRGECSGTASVVCLLNNFYPR= VYACEVTHQGLSSPVTK, z=+8, where 3 peptides are connected by 3 disulfide linkages
Though CID has been widely used as a disulfide mapping assay, EAD provided complementary information to CID in terms of peptide backbone sequence coverage. In this study, the sequence coverage of the disulfide-linked peptide was calculated at 32% for EAD and 18% for CID. This highly complex peptide, with 6 Cys residues connecting the 3 peptides by 3 disulfide links, exemplifies how EAD can achieve high confidence in peptide identification.
Scrambled disulfide linkages
Qualitative and quantitative assessment of scrambled disulfide linkages is a critical quality attribute of biologics. As such, several studies have investigated the impact of various sample preparation or storage conditions on the degree of scrambling of Cys residues in different IgG biotherapeutics.1,2,5 For example, Cys214 stabilizes the higher-order structure of adalimumab via the formation of an inter-chain disulfide linkage with Cys224 of the heavy chain4,5, and heating adalimumab at 70 °C for 30 min resulted in 100% scrambling at Cys214 residue of the light chain5. In general, interchain linkages are more susceptible to reduction, incomplete formation and shuffling than intrachain linkages.1
Figure 5. EAD (top right) and CID (bottom right) MS/MS spectra of the disulfide-linked peptide VYACEVTHQGLSSPVTK=SFNRGEC, Cys194=Cys214, a scrambled disulfide linkage from the light chain that is chromatographically resolved from its native form (left).
Figure 5 shows the peptide chromatogram, EAD and CID MS/MS spectra of VYACEVTHQGLSSPVTK=SFNRGEC (z=+5, 526.45 m/z), where a scrambled disulfide linkage is formed between Cys194 and Cys214 of the light chains. Compared with the expected disulfide linkage at Cys224:HC-Cys214 :LC, this scrambled-linked peptide is of low abundance, representing only 0.4% of disulfide linkages at Cys214 (Figure 5, left). Once again, EAD demonstrated high peptide backbone amino acid sequence coverage of 84% compared to CID coverage of 55%. This was achieved by generating fragment ions corresponding to the individual peptides after breaking the disulfide linkage.
EAD generated the signature fragment for Leu, z• 7-43 (w7, mass error=5.6 ppm, Figure 5), which further confirmed the presence of Leu residue and differentiated it from its isomer Ile. This additional level of information obtained through single injection of an EAD method enabled the confirmation of the identity of the connected peptides. This eliminates the possibility of misassignment, especially in the case of low-abundance scrambled disulfide linkages, thus minimizing the instrument time.
Free thiols
Free Cys have been reported as a cause of forming non-native disulfide bonds during manufacturing2, which can impact the protein’s 3-dimensional structure. Therefore, it is necessary to identify and monitor existing free thiols to avoid potential risks to drug safety. To accurately detect free thiols, all free Cys residues were capped with iodoacetamide before enzymatic digestion, leading to a +57 Da mass increase per alkylation. Low levels of free thiols were detected at different Cys residues of adalimumab. Figure 6 illustrates an example from peptide SGTASVVCLLNNFYPR z=+3, where carbamidomethylation was detected at Cys134. Both EAD and CID methods provided highly informative spectra with sequence coverage of 93% and 73%, respectively, thereby confirming the existence of a free thiol. Furthermore, the EAD spectrum offers additional capability to confirm the presence of 2 Leu residues through the detection of z• 7-43 (w7) and z• 8-43 (w8) signature fragments with mass error= 0.14 and -1.52 ppm, respectively.
Figure 6. EAD (Top) and CID (bottom) MS/MS spectra of the peptide SGTASVVCLLNNFYPR, where a free thiol was capped using iodoacetamide at Cys 134
Conclusion
- Confident sequence and disulfide linkage confirmation of long disulfide-linked peptides were achieved with EAD
- For peptides that contain 2 or more disulfide bonds, EAD generated rich fragments allowing reliable confirmation of individual peptides involved in the disulfides
- EAD spectra demonstrate superior fragmentation coverage on disulfide-linked peptides compared to CID
- Free thiols were detected at low levels with high confidence using both EAD and CID
- EAD produces signature ions to facilitate isomer differentiation, like Asp and isoAsp, Leu and Ile
- Biologics Explorer software facilitates automatic, state-of-the-art data processing for both routine and advanced characterization of complex linkages in biotherapeutics and standard monoclonal antibodies in a reproducibl e manner
References
- Coghlan J et al. (2022) Streamlining the characterization of disulfide bond shuffling and protein degradation in IgG1 biopharmaceuticals under native and stressed conditions, Front. Bioeng. Biotechnol. 10, 862456.
- Lakbub J et al. (2018) Recent mass spectrometry-based techniques and considerations for disulfide bond characterization in proteins, Anal. Bioanal. Chem. 410, 2467-2484.
- Huang L et al. (2019) Complete mapping of disulfide linkages for etanercept products by multi-enzyme digestion coupled with LC-MS/MS using multi-fragmentations including CID and ETD, J Food Drug Anal. 27.2, 531-541.
- Kwon J et al. (2021) Physicochemical and biological similarity assessment of LBAL, a biosimilar to adalimumab reference product (Humira®). Anim. Cells Syst. 25.3, 182- 194.
- Resemann A et al. (2018) Rapid, automated characterization of disulfide bond scrambling and IgG2 isoform determination, MAbs. 10.8,1200-1213.
 Click to enlarge
Click to enlarge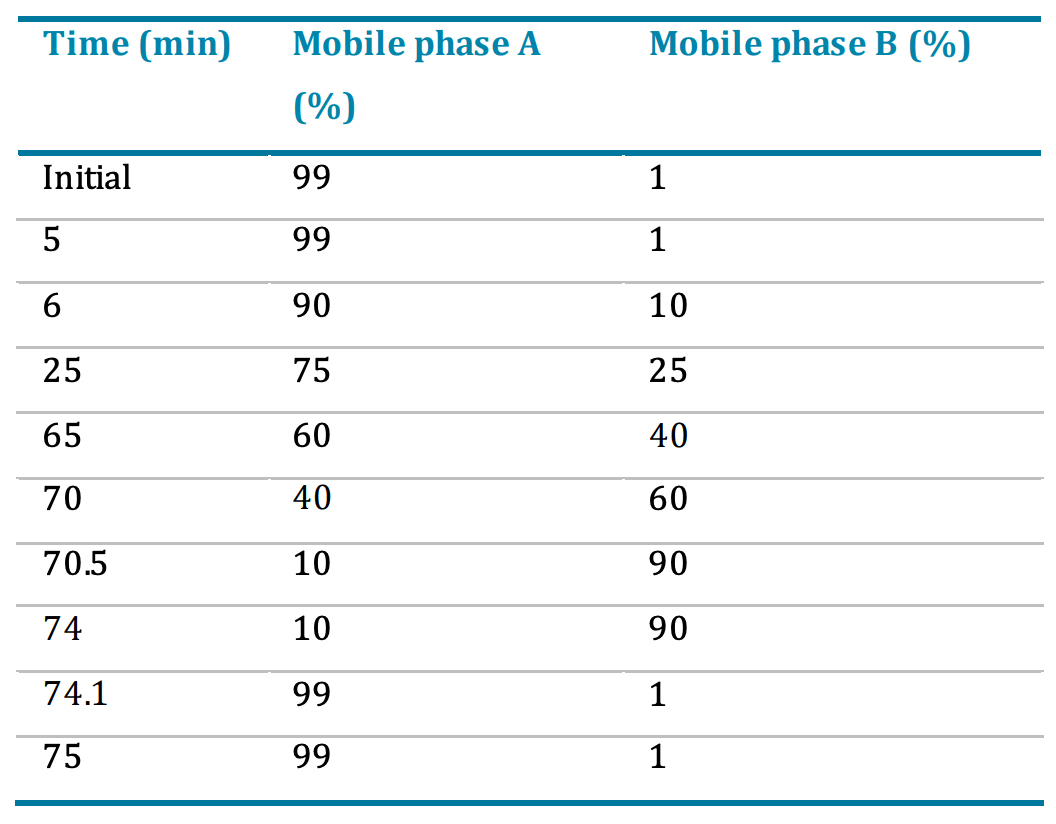 Click to enlarge
Click to enlarge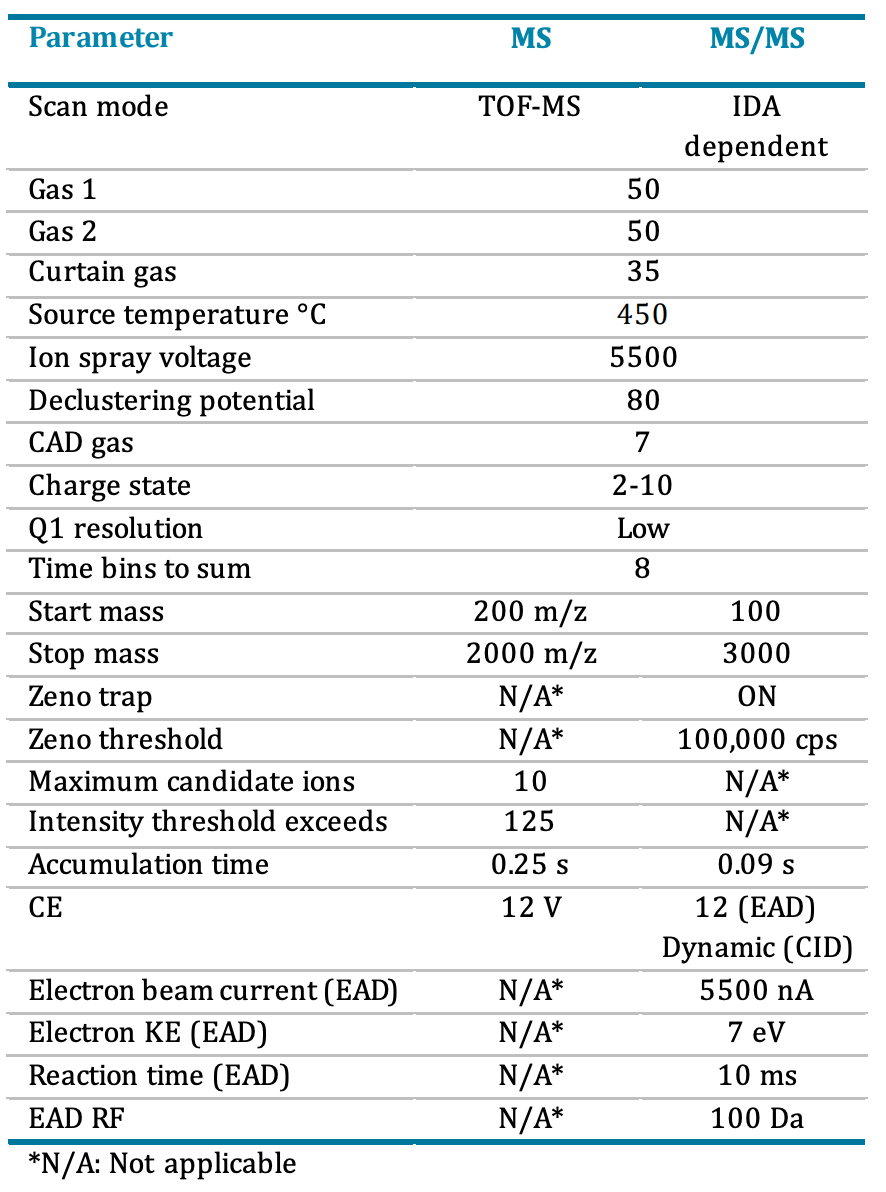 Click to enlarge
Click to enlarge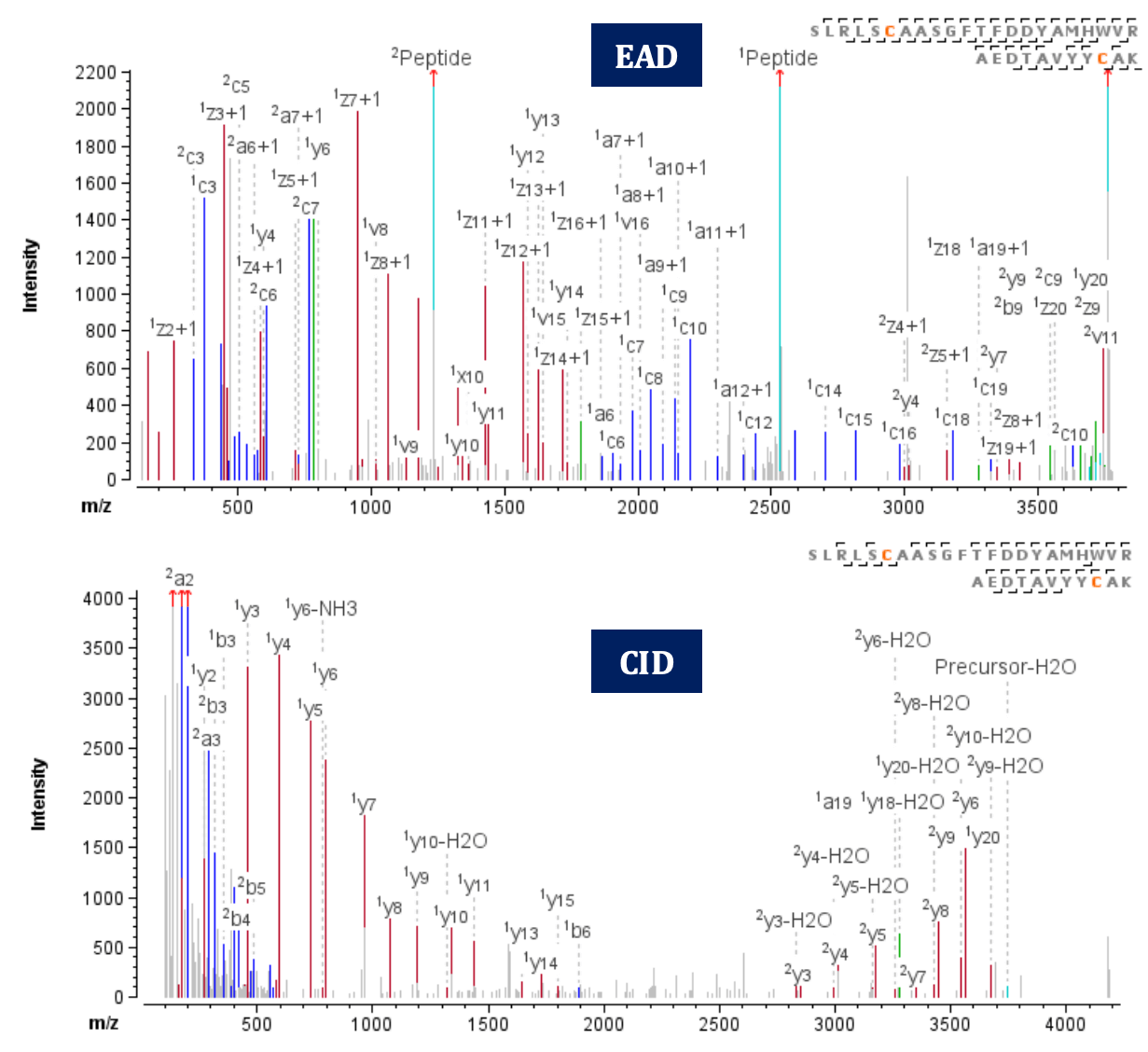 Click to enlarge
Click to enlarge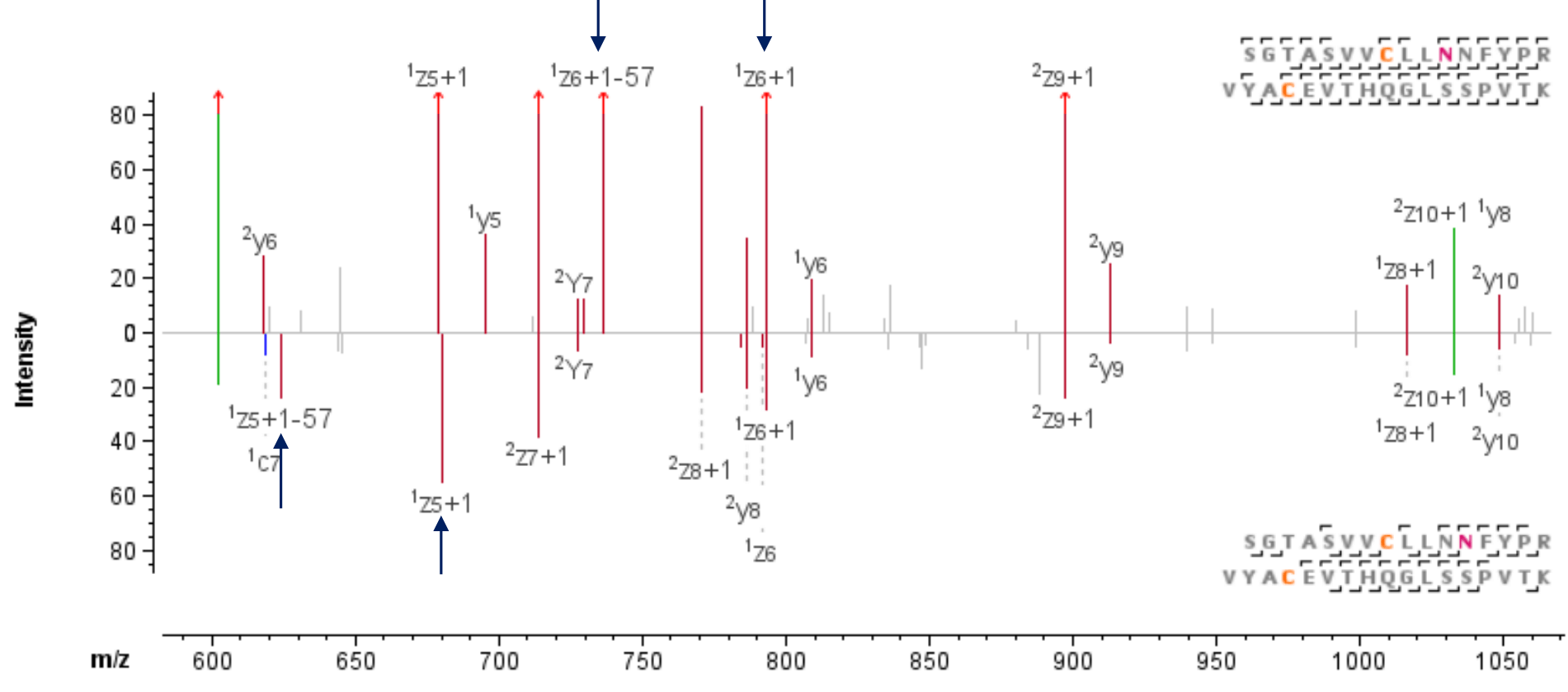 Click to enlarge
Click to enlarge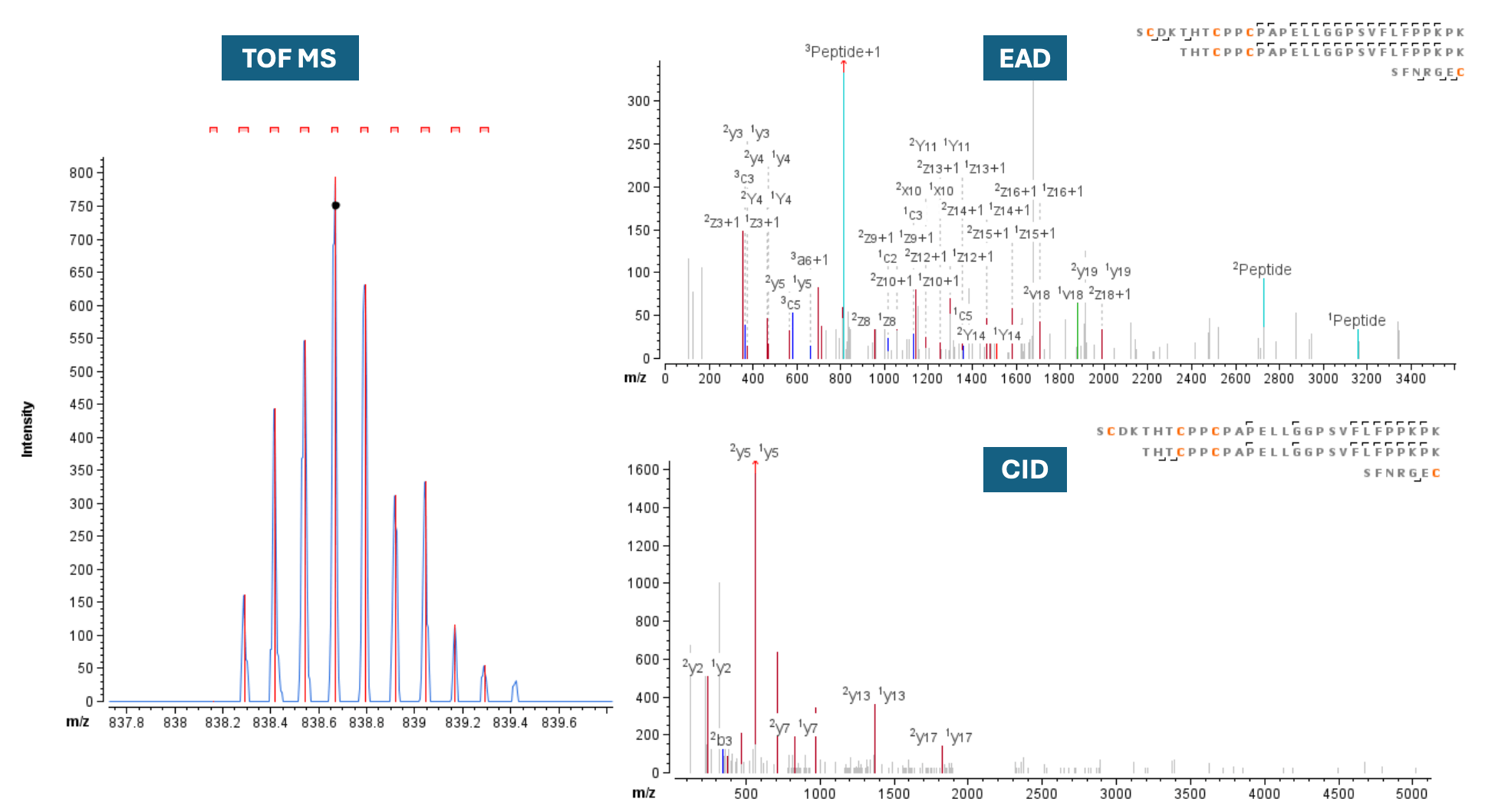 Click to enlarge
Click to enlarge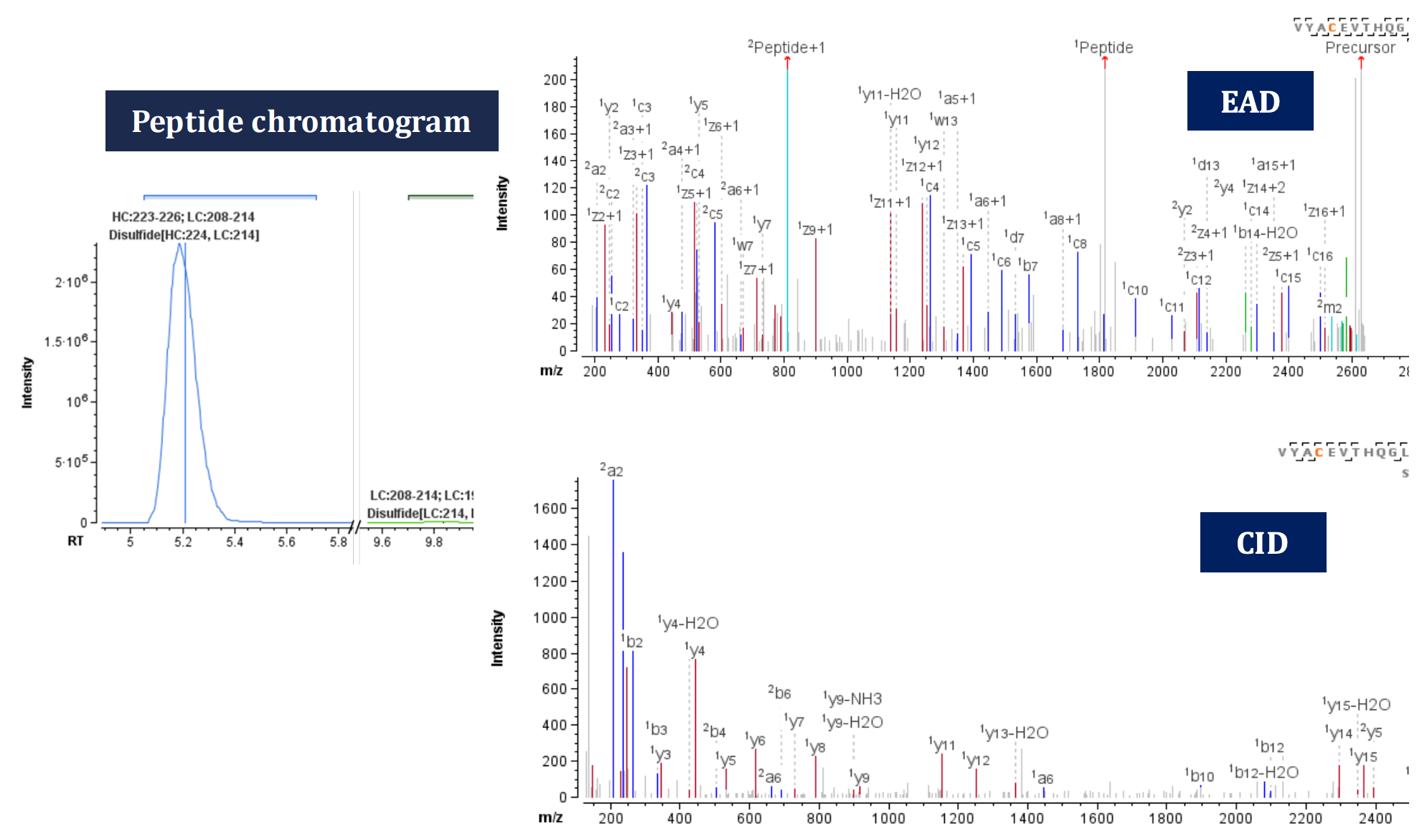 Click to enlarge
Click to enlarge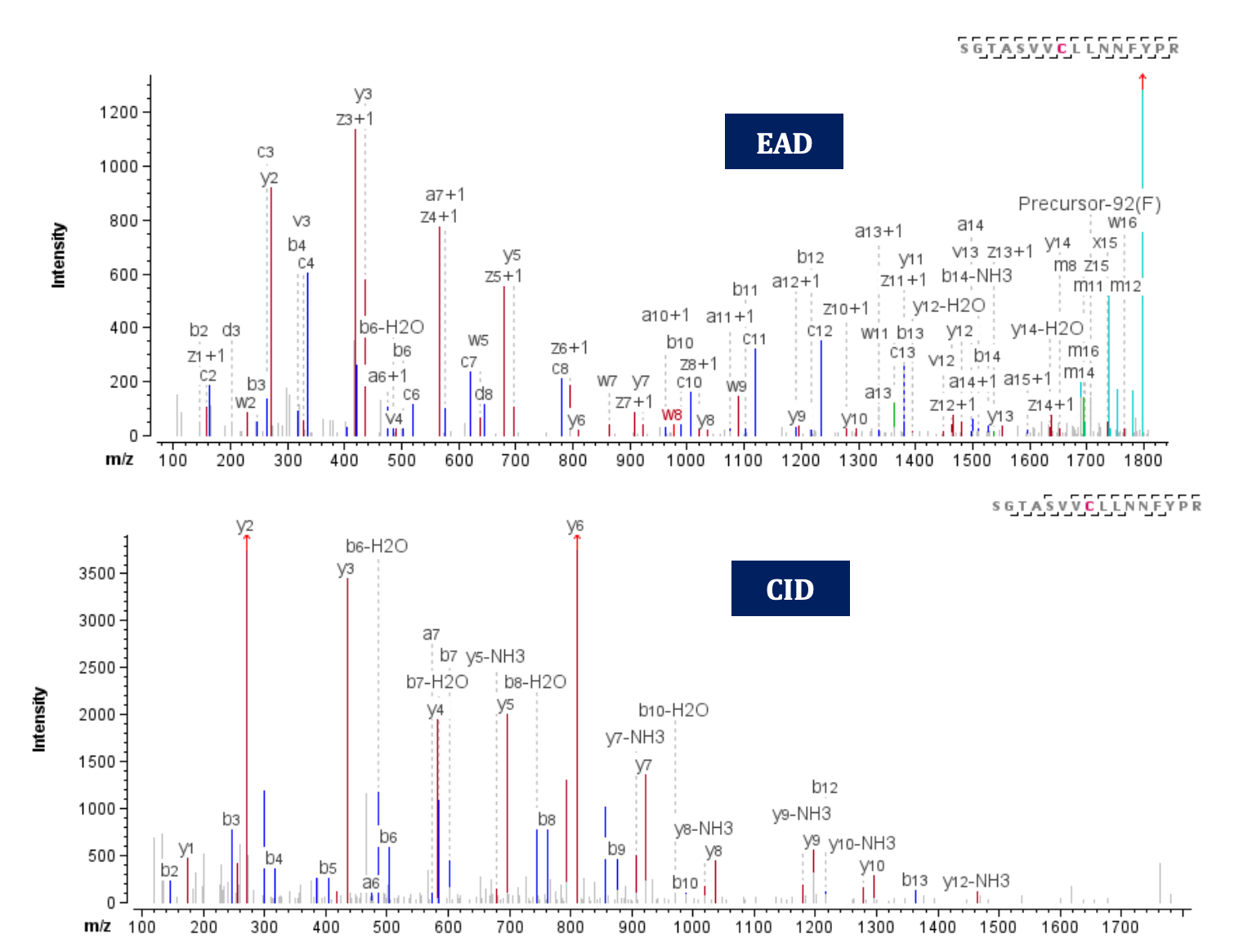 Click to enlarge
Click to enlarge