Abstract
This technical note demonstrates a novel integrated workflow using the Intabio ZT system. This innovative system offers direct chip-based integration of imaged capillary isoelectric focusing (icIEF) with mass spectrometry (MS), which affords confident characterization of proteins and reliable quantitation of charge variants by icIEF-UV/MS. Further, the streamlined icIEF-UV/MS platform significantly reduces the time to results from weeks or longer to approximately 1 hour for routine samples, compared to ion exchange chromatography (IEX) with fraction collection.
Introduction
Recombinant monoclonal antibodies (mAbs) have become an important class of biotherapeutics to treat a wide variety of diseases due to their high specificity, efficacy and flexibility.1 During the manufacturing process, heterogeneity of the mAbs could occur due to enzymatic cleavage and chemical posttranslational modifications (PTM).2 Many of these PTMs, including deamidation, C-terminal lysine truncation, glycation and sialylation, change mAb charge (and, thus, isoelectric point or pI).3 Characterizing the charge heterogeneity of mAbs is essential for critical quality attribute (CQA) assessment to ensure drug safety, efficacy and potency.4 In addition, process changes can happen during manufacturing, which poses a significant concern for biopharma therapeutic development. Process changes can result in PTMs that impact product quality.
cIEF offers a high-resolution separation of protein charge variants, including compounds with similar pI values.3 Therefore, cIEF is commonly used to monitor CQAs and product quality attributes (PQA). However, how to directly and rapidly identify and characterize different PTMs from these charge variant peaks in a single assay using iciEF-UV/MS data has been an unsolved problem for decades.
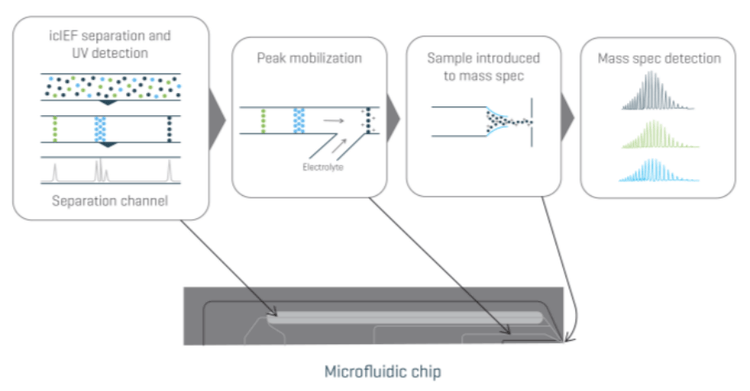
The Intabio ZT system aids in solving this challenge. It directly integrates icIEF separation with a ZenoTOF 7600 system, enabling direct coupling of icIEF charge variant analysis, UV quantitation and high-resolution mass spectrometry to identify intact proteins.
The UV results provide relative quantitation of the charge variant peaks. After focusing, separated charge variants are chemically mobilized and introduced to the MS through an on-chip electrospray ionization emitter for analysis without compromising separation efficiency and resolution. The icIEF-UV profile is correlated to the MS base peak electropherogram (BPE) for peak assignment and annotation.
The Intabio ZT system in combination with ZenoTOF 7600 system confidently identified numerous attributes, including deamidation and low-abundance glycation events. The total turnaround time for routine samples is approximately 1 hour, which offers significant time savings compared to the conventional LC IEX workflow and fraction collection followed by MS identification. Since all data are collected in a single MS and optical analysis, data integrity is superior to fraction collection for GLP considerations.
Key features
- Seamless identification of charge variants with a microfluidic chip-based integrated icIEF-UV/MS technology. The Intabio ZT system is exclusively coupled to the ZenoTOF 7600 system
- Platform offers both rapid multi-attribute monitoring of intact biotherapeutics and comprehensive characterization by icIEFUV/MS
- The 30-min sample analysis is significantly faster than conventional cIEF and IEX workflows requiring fractionation for the following identification.
- icIEF separation and UV quantitation correlate well with standard icIEF techniques
- Streamlined, intuitive data analysis software for rapid reporting and result sharing
Methods
Equipment: Intabio ZT system (SCIEX) and Intabio ZT cartridge (SCIEX, P/N 5088248) were used for the separation of NISTmAb and its charge variants. MS detection was performed on the ZenoTOF 7600 system (SCIEX, P/N 5080337) equipped with OptiFlow interface components (SCIEX, P/N 5084645).
Chemicals and reagents: The Intabio system – Electrolyte and Mobilizer kit (P/N 5088205) was used for anolyte, catholyte and mobilizer. Anolyte and mobilizer were used undiluted. The stock catholyte solution was 1% and diluted to 0.25% for use in the reagent drawer. The stock anolyte is 1% formic acid and catholyte is 1% diethylamine. The mobilizer is composed of 25% acetic acid, 25% acetonitrile and 50% water.
A 500mM cathodic spacer solution containing free base Larginine (Arg) (purity ≥ 98.5%, Sigma-Aldrich, P/N A8094-25G) was prepared by dissolving 0.870 mg of Arg powder into 10 mL of Milli-Q water. The electrolytes and cathodic spacer solutions were stored at room temperature. pI markers (CanPeptide) were individually dissolved in Milli-Q water at 5 mg/mL.
Prior to icIEF-UV/MS analysis, NISTmAb was desalted with a Zeba Spin Desalting Columns, 7K MWCO, 0.5 mL(Thermo Fisher Scientific, P/N 89882).
icIEF-UV/MS analysis: MS signal was optimized before sample analysis using a solution containing 100 µg/mL NISTmAb in mobilizer solution. The NISTmAb solution was infused through the Intabio ZT cartridge with 100-150 mbar of pressure, and electrospray was achieved into a ZenoTOF 7600 system with a tip voltage between 5000 and 5500 Volts. Nitrogen gas was applied between 60 to 75 psi to the on-chip integrated nebulization channels coplanar with the tip of the icIEF-UV/MS chip to achieve stable and robust ionization and sample entry to the MS.
Samples containing 250 µg/mL NISTmAb, 10 mM arginine, 1% Pharmalyte 3 to 10 (Cytiva, PN 17045601), 2.5% Pharmalyte 8 to 10.5 (Cytiva, PN 17045501), and 6.25 µg/mL peptide pI markers were vortexed and then degassed by centrifugation at 3900 rcf.
The streamlined process of icIEF-UV/MS analysis is illustrated in Figure 1. First, 70 psi of nitrogen was supplied to the internal nebulization channel within the Intabio ZT cartridge during the loading, focusing, and mobilization cycles of the icIEF-UV/MS separation process. After priming the channels within the Intabio ZT cartridge with either electrolyte solution or water, 50 µL of the sample was automatically loaded onto the separation channel using the autosampler. The icIEF separation was achieved using the parameters shown in Table 1. Absorbance measurements were collected at 1 Hz throughout the focusing and mobilization steps. The samples were introduced into the ZenoTOF 7600 system by a metered 2 µL/min flow of chemical mobilizer, and the data was acquired using parameters shown in Table 2.
Results and discussion
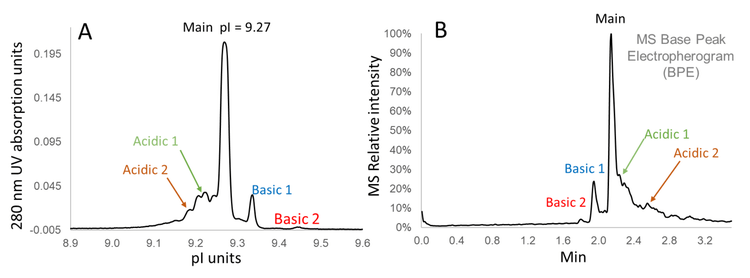
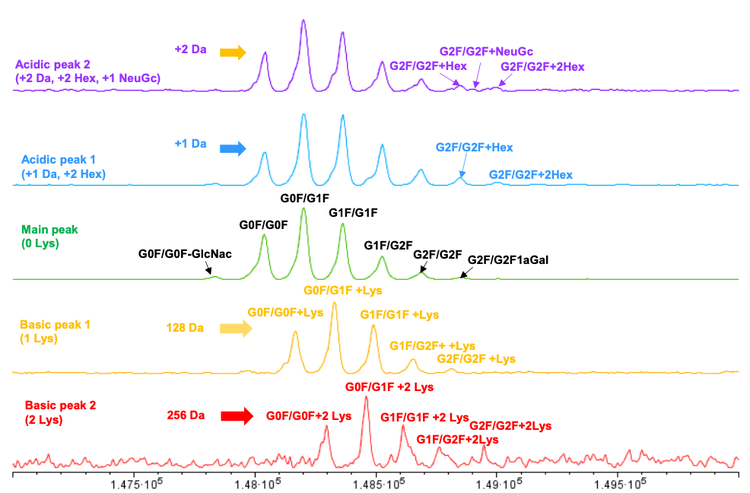
Figure 2 shows a separation profile of the charge variants of NISTmAb acquired with the Intabio ZT system coupled to ZenoTOF 7600 system. Panel A shows a 280-nm UV absorbance profile of NISTmAb charge variants separated by icIEF. Panel B is a MS BPE of the corresponding peaks after mobilization, electrospray and detection by MS. The separation profiles generated by icIEF-UV (once inverted) and icIEF-MS on the Intabio ZT system and ZenoTOF 7600 system, respectively, are similar. icIEF-UV profile shows separated peaks across the icIEF separation channel, whereas the MS data is acquired based on the time the charge variants are mobilized and electrosprayed into the mass spectrometer. Basic peaks of the icIEF-UV profile with high pI are introduced first into the MS system for analysis and therefore appear at earlier time points (left side of the MS BPE). The MS BPE shows that all the peaks observed from icIEF-UV profile were also detected by the MS system without compromising separation resolution. This result indicates that the separation efficiency of charge variants was well maintained after chemical mobilization. The entire focusing and separation only took 15 min, demonstrating the capability of high-throughput characterization of charge variants.
A detailed analysis of the charge variant peaks was performed on intact NISTmAb. Figure 3 shows the intensity normalized deconvoluted mass spectra of 5 charge variants from the NISTmAb sample. The basic peaks 2 and 1 charge variants show +256 and +128 Da mass increases compared to the main peak, which are respectively identified as C-terminal lysine variants.
Additional peaks above G2F/G2F at +162 Da intervals are observed in acidic peaks 1 and 2, identified as putative glycation. Masses that correspond to glycans with NeuGc sialic acid are observed in acidic peaks 2. A +1 Da mass shift was observed in acidic peak 1 and a +2 Da mass shift was observed in acidic peak 2 for proteoforms with G0F/G0F, indicating a high abundance of proteoforms with1 deamidation or 2 deamidation events in these 2 charge variants. Due to the excellent charge heterogeneity separation on the Intabio ZT system, neutral proteoforms, such as the glycosylation series, including lowabundance glycans such as G0F/G0F-GlcNAc, are readily measured by icIEF-UV/MS. In a follow-up technical note, we confirm the existence of glycation by performing the icIEFUV/MS analysis on a deglycosylated NISTmAb using the Intabio ZT system.
Conclusion
- The Intabio ZT system demonstrates a streamlined workflow to separate charge variants by icIEF and identify them with ZenoTOF 7600 system
- A unified icIEF-UV/MS workflow offers high sensitivity to detect low-abundant charge variants that might impact product quality
- The separation efficiency and resolution of icIEF are wellmaintained after mobilization, enabling highly confident identification of CQAs by icIEF-UV/MS, such as C-terminal lysine variants, deamidation and glycation (Hex)
- The Intabio ZT system is a commercially available platform that offers workflow combining icIEF separation, UV quantitiation and MS-based identification.
References
- Mullard A. FDA approves 100th monoclonal antibody product. Nat Rev Drug Discov. 2021 Jul;20(7):491-495.
- Ostrowski M. Raplid multi-attribute characterization of intact bispecific antibodies by a microfluidic chip-based integrated icIEF-MS technology. Electrophoresis. 2023, 44:378-386
- Mack S, Arnold D, Bogdan G, et al. A novel microchipbased imaged CIEF-MS system for comprehensive characterization and identification of biopharmaceutical charge variants. Electrophoresis. 2019, 40: 3084-3091
- Houde D, Peng Y, Berkowitz SA, Engen JR. Posttranslational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics. 2010 Aug;9(8):1716-28.

