Introduction
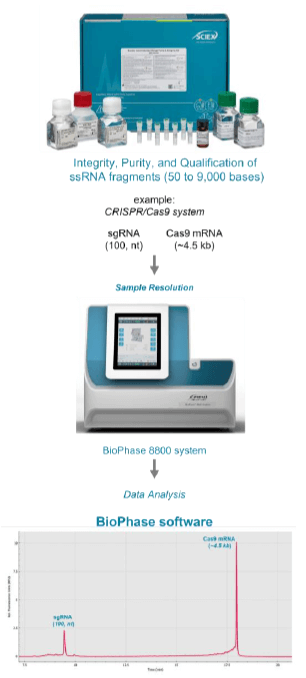
RNA vaccine and CRISPR (clustered regularly interspaced short palindromic repeats) reagents manufacturers are often challenged to accurately characterize and quantify differently sized RNA molecules, impurities, and degraded RNA species present in vaccines or personalized medicinal products 1,2. To help overcome these challenges, in this technical note, we present an analytical kit-based solution for the characterization of both RNA integrity in the final product and RNA fragmentation. Using the multi-capillary electrophoresis platform, we demonstrate an efficient and timesaving workflow to assess various critical quality attributes (CQAs) for potential mRNA vaccines and CRISPR reagents (Figure 1). Excellent reproducibility was obtained for the purity content of the main product of CRISPR/Cas9 gene editing system with a %CV of <2%. These results demonstrate the ability of the RNA 9000 Purity & Integrity Kit to separate single-stranded RNA products in the range of 50 to 9,000 bases.
Key features
- Ability to acquire high-resolution data for RNA fragments ranging from 50 to 9,000 bases using the RNA 9000 Purity & Integrity Kit
- Excellent %CV, < 2%, for the corrected peak area% of the main product of the Cas9 mRNA component of the CRISPR/Cas9 gene editing system
- Ready-to-use, validated kit and methods enable efficient RNA fragment characterization using the BioPhase 8800 system
RNA vaccines and CRISPR-based gene editing are two recent breakthroughs used to combat SARS CoV-2 or potentially treat many genetic diseases, respectively.1,2 The safety and efficacy of RNA vaccines and CRISPR-based gene editing products highly depend on the stability and purity of the nucleic acid in the final product.
In this technical note, we used the RNA 9000 Purity & Integrity Kit with the BioPhase 8800 system to characterize a chemically modified sgRNA targeting the human HPRT1 gene (100 bases) and the Cas9 mRNA (~4.5 kb). The integrity and purity of the sgRNA and Cas9 mRNA were assessed using capillary gel electrophoresis with laser-induced fluorescence (CGE-LIF).
This technical note highlights the efficiency of our optimized electro kinetic injection-based separation method developed for the RNA 9000 Purity & Integrity Kit and the BioPhase 8800 system.
Methods
Materials: The RNA 9000 Purity & Integrity Kit (PN: C48231) was from SCIEX (Framingham, MA) and contained the Nucleic Acid Extended Range Gel, SYBRTM Green II RNA Gel Stain*, Acid Wash (regenerating solution), CE-grade water, and the ssRNA Ladder (50 to 9,000 bases). Pre-assembled BioPhase BFS Capillary Cartridge (8 capillaries, 30 cm total length, PN: 5080121), the disposable BioPhase sample and reagent plates (PN: 5080311), and the sample loading solution (SLS, PN: 608082) were from SCIEX (Framingham, MA).
The CleanCap Cas9 mRNA (5moU) (PN: L-7206) was obtained from TriLink Biotechnologies (San Diego, CA). The Cas9 mRNA had an expected size of 4,521 bases, as characterized by agarose gel mobility and was shipped at a concentration of 1.0 mg/mL.
A proprietary crRNA sequence (20 bases) targeting the human HPRT1 gene (NM-000194) was fused by synthesis to the tracrRNA sequence (80 bases) at 10 nmol by Integrated DNA Technologies (IDT; Coralville, IA) to generate a long, sgRNA (100 nucleotides; 32,458.9 g/mole). This Alt-R CRISPR-Cas9 sgRNA product contained chemical modifications to increase the sgRNA stability, potency, and resistance against nuclease activity.4
Instrumentation: The multi-capillary BioPhase 8800 system equipped with a laser induced fluorescence (LIF) detector was from SCIEX (Framingham, MA).
Sample dilution and storage: The Cas9 mRNA was stored at -80°C until the time of analysis. Working samples were diluted with CE-grade water to 100 ng/μL in 20 μL aliquots from the main stock solution (1 mg/mL). The lyophilized sgRNA was resuspended at 3.246 μg/μL (100 μM), using a Tris-EDTA pH 7.5 solution containing 10mM Tris, 0.1mM EDTA, aliquoted into small volumes and stored at -80°C until the time of analysis. A 100 μL aliquot of sgRNA working solution was prepared by diluting the stock solution (3.25 μg/μL) to 100 ng/μL using CE-grade water.
Cas9 mRNA sample preparation: A 0.4 ng/μL sample solution was prepared for BioPhase 8800 analysis. To prepare the 0.4 ng/μL Cas9 mRNA analysis sample, first the Cas9 mRNA 100 ng/μL working solution was diluted tenfold (1 to 10) using CE-grade water, followed by mixing 2 μL of the tenfold Cas9 mRNA dilution with 48 μL of a 50:50 CE-grade water/SLS solution to yield a total volume of 50 μL.
sgRNA sample preparation: A 25 ng/μL sample solution was prepared for BioPhase 8800 analysis. To make the 25 ng/μL sgRNA analysis sample, 12.5 μL from the sgRNA working solution (100 ng/μL) were mixed with 37.5 μL of a 50:50 CE-grade water/SLS solution to yield a total volume of 50 μL.
Cas9 mRNA and sgRNA co-analysis: For the co-analysis of the Cas9 mRNA and the sgRNA, 2 μL of the tenfold Cas9 mRNA dilution and 12.5 μL from the sgRNA working solution (100 ng/μL) were mixed with 35.5 μL of a 50:50 CE-grade water/SLS solution to yield a total volume of 50 μL.
Cas9 mRNA sample load analysis: A 2 ng/μL, a 1.2 ng/μL, and 0.4 ng/μL sample solutions were prepared for BioPhase 8800 analysis. To make the 2 ng/μL Cas9 mRNA analysis sample, a 1 μL from the 100 ng/μL working solution was mixed with 49 μL of a 50:50 CE-grade water/SLS solution to yield a total volume of 50 μL. To prepare the 1.2 ng/μL Cas9 mRNA analysis sample, first the Cas9 mRNA 100 ng/μL working solution was diluted tenfold (1 to 10) using CE-grade water, followed by mixing 6 μL of the tenfold Cas9 mRNA dilution with 44 μL of a 50:50 CE-grade water/SLS solution to yield a total volume of 50 μL. The 0.4 ng/μL Cas9 mRNA analysis sample was prepared by mixing 2 μL of the tenfold Cas9 mRNA dilution with 48 μL of a 50:50 CE-grade water/SLS solution to yield a total volume of 50 μL.
After the above mixtures were prepared, they were denatured with heat using a thermal cycler at 70°C for 5 minutes, then immediately placed on ice until time of injection for CE analysis using the BioPhase 8800.
ssRNA ladder preparation: The ssRNA ladder was prepared according to the RNA 9000 Purity & Integrity Kit Application Guide. A 50 μL sample was denatured with heat using a thermal cycler at 70°C for 5 minutes and immediately placed on ice until time of injection for CE analysis. The number of prepared ssRNA ladder samples depended on the number of replicates needed.
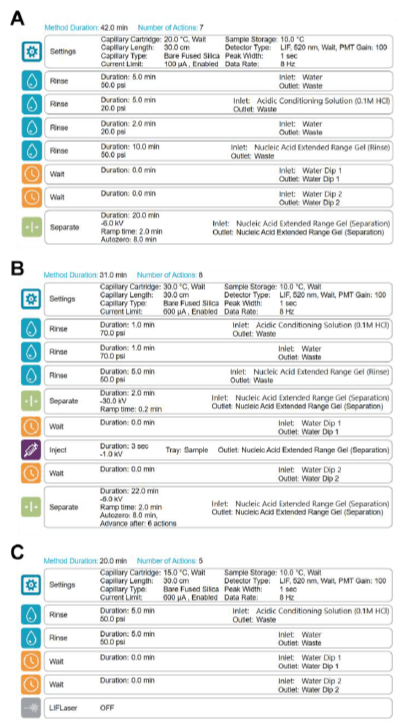
CE separation method: Figure 2 illustrates the BioPhase 8800 system operation sequence for the conditioning (panel A), separation (panel B) and shut down (panel C) methods used for this study. For a detailed sample preparation workflow and BioPhase 8800 operation refer to the RNA 9000 Purity & Integrity Kit Application Guide.5
Data processing: Unless indicated, peak analysis was performed by applying a 0.75% positive threshold with suspended integration from 0 to 8 minutes. The BioPhase software version 1.0 was used to calculate corrected peak area (CPA) and corrected peak area % (CPA%) for the main product, nucleic acid impurities, and higher molecular weight (HMW) products.
Results and discussion
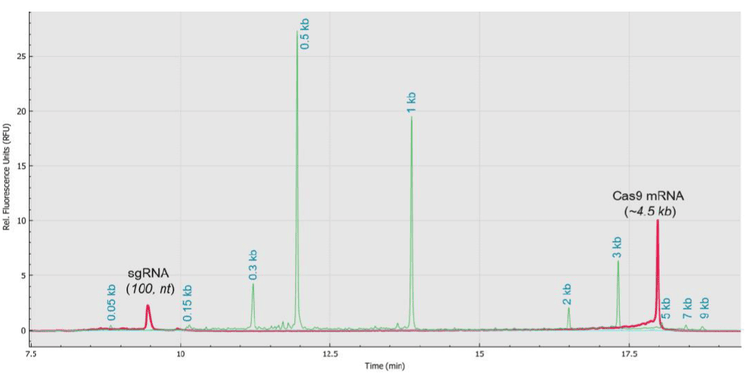
The objective of this technical note was to demonstrate the ability of the RNA 9000 Purity & Integrity Kit from SCIEX to differentiate single-stranded RNA fragments of various sizes in the range of 50 to 9,000 bases with high-resolution. Figure 4 illustrates the workflow for simultaneous analysis the small (100 bases) and large (~4.5 kb) RNA fragments from the CRISPR/Cas9 system to showcase the robustness and timesaving sample analysis using the RNA 9000 Purity & Integrity Kit with the BioPhase 8800 system. Once samples were analyzed, data assessment was performed with the BioPhase software version 1.0. The representative electropherogram shown at the bottom of Figure 4, which was acquired on the BioPhase 8800 system demonstrates the simultaneous profiling of small and large RNA products used for gene editing applications.
An additional analytical asset provided in the RNA 9000 Purity & Integrity Kit is the ssRNA ladder that contains RNA fragments with lengths of 50, 150, 300, 500,1,000, 2,000, 3,000, 5,000, 7,000 and 9,000 bases. These RNA size markers, and shown in the following sections, provided the built-in technology for estimating the size of the main products and impurities.
CRISPR/Cas9 nucleic acid profiling, quantification and repeatability using the multi-capillary BioPhase 8800 system
HPRT1 sgRNA characterization
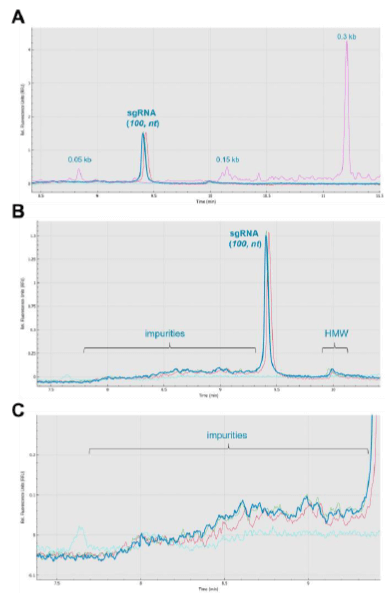
Figure 5A shows the overlay of 3 injections (red, green, and blue traces) of a sgRNA sample (100 bases). The pink trace shows the ssRNA ladder and corresponding sizes. The background signal acquired from an injection of water is indicated by the aqua trace. The sgRNA peaks appeared between the 50 bases and 150 bases ssRNA molecular markers. Figure 5B shows the overlay of the triplicate injections without including the RNA ladder. This analysis displayed a better visualization of the RNA fragment impurities and the RNA HMW products. Figure 5C shows the zoomed-in view of the region in front of the full-length sgRNA peak. The RNA fragmentation or impurities in this region were most likely from incomplete sgRNA synthesis such as n-1 species.
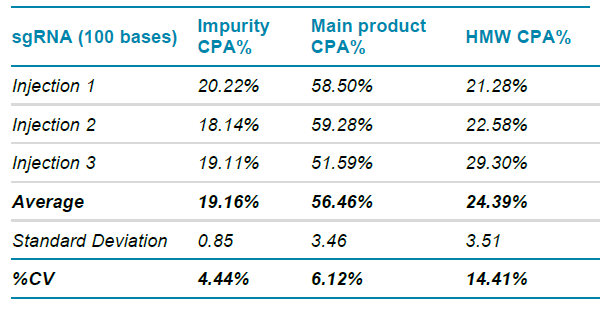
Table 1 shows the repeatability injections CPA% measurements from the same capillary attained on the multi-capillary BioPhase 8800 system for the nucleic acid products shown in Figure 5. Measurements were based on 3 consecutive injections of a sample preparation, as described in Methods. The nucleic acid impurities, main sgRNA product, and HMW content accounted for average CPA% values of 19.16%, 56.46%, and 24.39%, respectively. The relatively low standard deviation calculations for these measurements demonstrate the high repeatability performance of the RNA 9000 Purity & Integrity Kit on the BioPhase 8800 system. Demonstrating good injection repeatability with a calculated %CV for the main product of the sgRNA of 6.12%, suggesting the application of the BioPhase 8800 and the RNA Purity & Integrity Kit for assessment of sgRNA products.
Cas9 mRNA characterization
Figure 6A shows the overlay of 3 injections (red, green, and blue traces) of a Cas9 mRNA sample (~4.5 kb). The pink trace shows the ssRNA ladder and corresponding sizes. The Cas9 mRNA peaks appeared between the 3 kb and 5 kb ssRNA size markers.
Figure 6B shows the overlay of the triplicate injections without including the RNA ladder. This analysis displayed a better visualization of the RNA fragment impurities and the RNA HMW products. Figure 6C shows the zoomed-in view of the region in front of the full-length Cas9 mRNA peak. This analysis revealed the presence of fragmentation or impurities in this Cas9 mRNA sample, possibly from degradation of the full-length Cas9 mRNA.
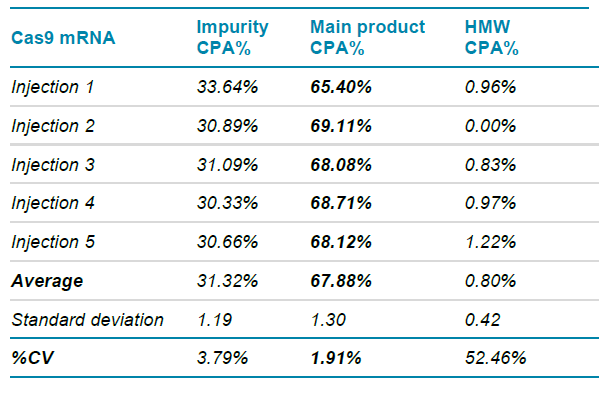
Table 2 shows the repeatability CPA% measurements attained on the BioPhase 8800 system for the nucleic acid products shown in Figure 6. Measurements were based on 5 consecutive injections of a sample preparation, as described in Methods (first three injections shown in Figure 6 as representative injections). The nucleic acid impurities, main Cas9 mRNA product, and HMW content accounted for average CPA% values of 31.32%, 67.88%, and 0.80%, respectively. The relatively low standard deviation calculations for these measurements demonstrate the high repeatability performance of the RNA 9000 Purity & Integrity Kit on the BioPhase 8800 system. In summary, we calculated a %CV for the main product of the sgRNA sample to be an outstanding 1.91%, suggesting the application of the BioPhase 8800 and the RNA Purity & Integrity Kit for assessment of the Cas9 mRNA component of the CRISPR/Cas9 gene editing system.
Simultaneous Cas9 mRNA and sgRNA characterization
Figure 7A shows the overlay of 3 injections (red, green, and blue traces) of a sample containing Cas9 mRNA and the HPRT1 sgRNA. The pink trace shows the ssRNA ladder and corresponding sizes. The background signal acquired from an injection of water is indicated by the aqua trace. Consistent with our previous results, the Cas9 mRNA peaks appeared between the 3 kb and the 5 kb ssRNA molecular markers. The sgRNA peaks also appeared between the 50 bases and 150 bases ssRNA size markers as shown by the RNA profiling of the sgRNA alone. These results suggest that mixing the RNA components of the CRISPR/Cas 9 gene editing system for purity and integrity co-characterization does not alter the structure or mobility of the Cas9 mRNA or the sgRNA during CGE-LIF analysis. However, prior to performing co-analysis of these RNA molecules, a comprehensive evaluation of these CRISPR/Cas9 gene editing system components should be performed as demonstrated in the sections analyzing the Cas9 mRNA and the sgRNA molecules on their own and without mixing these RNA components. A main advantage of the RNA 9000 Purity & Integrity Kit is the ability to resolve single stranded RNA fragments or products in the range of 50 bases to 9,000 kb, in this case the Cas9 mRNA and the sgRNA are excellent examples for the analytical applications of this Kit to accelerate the research and development pipeline or product release of RNA based therapeutics or vaccines.
Figures 7B through 7D show the individual RNA profiling of the overlayed injections as shown in Figure 7A. This extension into individual analysis of each injection from a sample containing mixed Cas9 mRNA and sgRNA demonstrates the reproducibility of CGE-LIF RNA profiling by using the BioPhase 8800 and the RNA 9000 Purity & Integrity Kit. Specifically, the red trace representing the mixed Cas9 mRNA and sgRNA sample, the green trace corresponding to the ssRNA ladder from the RNA 9000 Purity & Integrity Kit, and the aqua trace illustrating signal arising from water injection used as a negative control.
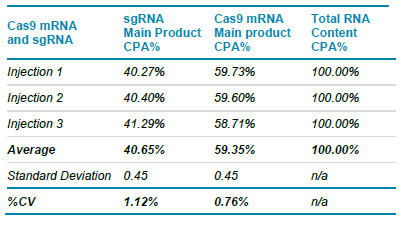
Table 3 summarizes the corrected peak area % of the Cas9 mRNA and the sgRNA mixed sample for each injection as shown in Figures 7B through 7D. This analysis involved setting up a 5.00% positive threshold to exclude RNA fragmentation or impurities in the sample mix. As a result, we obtained excellent reproducibility for the mixture containing 0.04 ng/uL of the Cas9 mRNA and 25 ng/uL of the sgRNA component. Specifically, an average Cas9 mRNA content of 59.35% and of the sgRNA of 40.65% were determine using the BioPhase software, respectively. A standard deviation value of 0.45 and %CV values of less than 2% were calculated based on this data set. Although, this analytical set up was not designed to reflect a specific stoichiometric relationship between the Cas9 mRNA and the sgRNA molecules, this analytical simulation demonstrates the applicability of the BioPhase 8800 system and the RNA 99000 Purity & Integrity Kit for monitoring or measuring pre-defined molecular ratios among differently sized single stranded RNA products in the range of 50 bases to 9,000 kb.
RNA sample concentration and RNA stability: Key variables for evaluating RNA products and assay development
RNA sample concentration and peak shape
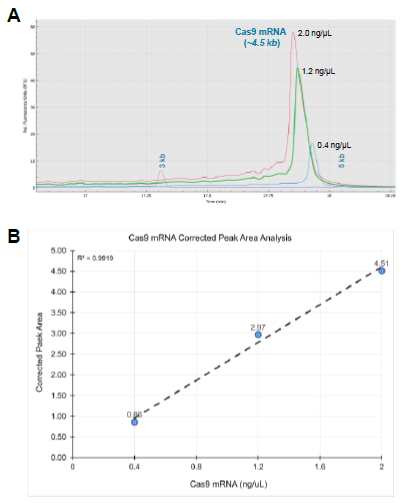
A powerful feature of the BioPhase 8800 is the accompanying software allowing in-depth peak analysis for single stranded RNA fragments. Among other peak measurements, peak symmetry can be automatically calculated to support the characterization of RNA samples used for CGE-LIF analysis. Specifically, we noticed that RNA concentration or amount of the same RNA molecule influences the peak symmetry. Figure 8A shows the overlay of the Cas9 mRNA molecule at 2.0 ng/uL (red trace), 1.2 ng/uL (green trace), and 0.4 ng/uL (blue trace). In contrast to the 2.0 ng/uL Cas9 mRNA sample, the 0.4 ng/uL Cas9 mRNA sample peak tailing seemed minimized. Specifically, we measured a peak symmetry of 1.45 for the 2.0 ng/uL Cas9 mRNA sample, a peak symmetry of 1.46 for the 1.2 ng/uL Cas9 mRNA sample, and a peak symmetry of 0.93 for the 0.4 ng/uL Cas9 mRNA sample. As a result, RNA sample concentration should be considered an important variable for assessing RNA stability and profiling. Figure 8B, shows a graphical analysis of the Cas9 mRNA samples and the corresponding corrected peak area values obtained by using the BioPhase software. Our analysis showed a linear trend with an R-squared value of 0.9919 for these representative injections. In summary, sample preparation techniques must use caution and interpretation of RNA peaks with varying peak fronting, peak tailing, peak ghosting, and assess whether the peaks are rounded or have a sharp at its crest.6 Controlling for peak variation or peak shapepattern could favor a robust assay development for final product qualification, release, or application of lifesaving technology.
Application of the RNA 9000 ladder for RNA stability studies
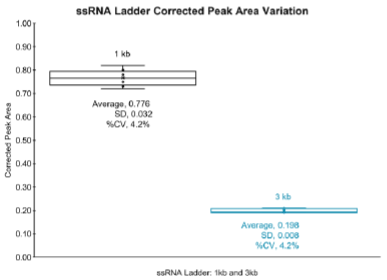
Due to naturally occurring RNA degradation events, because of physical or chemical processes, the integrity and purity profiling may vary for some RNA products. Nevertheless, and as previously cited, capillary gel electrophoresis is a frequently used technique that can give information about the size and the integrity of RNA products.7 As a result, in this technical note, we demonstrate the application of the ssRNA ladder included in the RNA 9000 Purity & Integrity Kit as a potential molecular marker for stability assessment of RNA products used for CGE-LIF analysis. Figure 9 shows the corrected peak area measurements based on a series of injections obtained from RNA 9000 ladder analysis. The CPAs for the 1 kb and 3 kb RNA size markers were graphed using a box and whisker plot to assess the injection variability of the RNA ladder during CGE-LIF analysis. Briefly, we calculated an average CPA of 0.78 for the 1 kb ssRNA marker with a %CV of less than 5%. The 3 kb ssRNA marker showed an average CPA of 0.20 with a %CV of less than 5%. These CPAs values obtained from a repeatability study of eight injections demonstrated the stability of the ssRNA markers, 1 kb and 3kb, respectively. These results suggest that the RNA 9000 ladder could serve as an analytical tool to assess the RNA stability of ssRNA products of varying sizes and different chemistries, as a result of varying nucleotide type content, or chemical modifications of RNA products.
Conclusion
- The RNA 9000 Purity & Integrity Kit provides a high-resolution separation solution that allows for impurity detection with excellent repeatability for small and large RNA products
- The RNA 9000 molecular weight ladder provides guidance for the qualification of differently sized RNA products
- Assay performance for the main expected product with %CVs <2% supports using CGE-LIF with the multiplexing capability of the BioPhase 8800 system for analytical characterization and large-scale manufacturing applications
References
- Damase, T. R., et. al., Front. Bioeng., Biotechnol. 2021. 9:628137.
- Tenchov R., et. al. ACS Nano, 2021, 15, 16982-17015.
- Eisenstein, M. Nature, 2022, 601, 658-661.
- Alt-R CRISPR-Cas9 sgRNAs. Integrated DNA Technologies. Technical Content Marketing.
- RNA 9000 Purity & Integrity Kit for the PA 800 Plus System Application Guide. SCIEX. 2022.
- Understanding chromatogram peaks – fronting, tailing, ghosting, and rounded explained. ChemTech International. 2020.
- Schoenmaker, L., et. al. International Journal of Pharmaceutics. 601.2021.120586

