Abstract
This technical note demonstrates a highly sensitive quantification workflow for surfactant proteins at a signature peptide level using a high-end triple quadrupole mass spectrometer. Enhanced sensitivity for quantification of surfactant proteins was achieved with the improved front-end technology to generate, capture and transmit more ions on the SCIEX 7500 system. Low-ng/mL lower limits of quantification (LLOQs) were achieved for the 4 surfactant proteins analyzed in human plasma.
Introduction
Despite the recent developments in LC-MS based protein biomarker quantification, sensitive multiplexed quantification of proteins remains a challenging step in the early stages of drug discovery and development. Biomarker targets such as surfactant proteins (SFTPs) are essential for facilitating and modulating the inflammatory response in the respiratory system. SFTP quantification in plasma is highly informative, as it enables the monitoring of lung health in an easily accessible matrix.
Quantification of protein biomarkers such as SFTPs is commonly performed using LC-MS techniques as it allows for the ability to multiplex. However, advanced sample pretreatment is required to extract protein biomarkers from matrix and sensitive analytical platforms are necessary to achieve quantification for low abundant targets. Target enrichment such as immunoprecipitation is often applied for quantification. However, it can be challenging and time consuming to develop an effective antibody particularly in the case of early drug discovery and development. Additionally, interferences such as anti-drug antibodies and low drug tolerance can further hinder immunocapture assay sensitivity.
Here, a novel antibody-free platform (ABFP) was applied for the enrichment of peptides in complex biological matrices. 1 ABFP employs a highly efficient chromatographic fractionation technique for signature peptide enrichment. Peptide analysis was performed using a microflow LC with trap-and-elute setup on the SCIEX 7500 system. Greater sampling efficiency of the microflow LC paired with the improved front-end technology on the SCIEX 7500 system substantially increased the overall sensitivity of the assay for SFTP biomarker quantification.
Key features of the protein biomarker quantification assay on the SCIEX 7500 system
- Accomplish sensitive multiplexed quantification of surfactant proteins in plasma at low-ng/mL level using the SCIEX 7500 system
- Achieve 2- to 5-fold improvement in LLOQ using the improved front-end technology on the SCIEX 7500 system compared with previous generation instrumentation
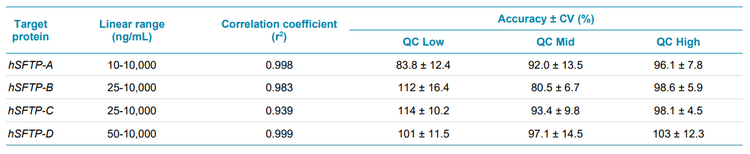
- Reach outstanding accuracy, precision and linearity for quantification of multiple protein targets in complex matrices
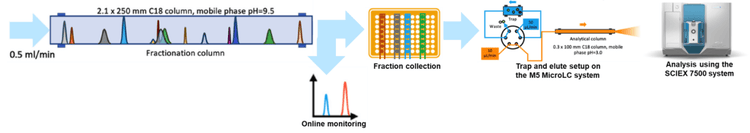
Methods
Sample preparation: Human SFTPs (hSFTPs) including hSFTP-A, hSFTP-B, hSFTP-C and hSFTP-D were enzymatically digested in human plasma. Details of the sample preparation protocol are provided in the manuscript.1
Chromatography: Sample fractionation was on BioZen NX-C18 Column (150 x 2.1 mm), using high pH (pH=9.5) mobile phase, Separation was performed on a M5 MicroLC system operated in a trap-and-elute mode. A Phenomenex Luna Omega polar trap column (0.5 × 10 mm, 5 µm, 100 Å) was used for analyte trapping at a flow rate of 50 µL/min. The trap column was operated at room temperature. Sample separation was performed on a Phenomenex Luna C18 (2) column (0.3 × 100 mm, 3 µm, 100 Å) at a flow rate of 10 µL/min. The column temperature was set to 50°C. Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% acetic acid in acetonitrile for both analyte trapping and separation.
Overview of the workflow for surfactant protein quantification assay
A total of 4 surfactant proteins with different physiochemical properties were analyzed in plasma. This included hSFTP-A, hSFTP-B, hSFTP-C and hSFTP-B. The ABFP technique was applied to enrich the signature peptides. A concentration range between 10 ng/mL and 10,000 ng/mL was measured. A total of 2 replicates were assessed for each calibration point. Analysis was performed on the SCIEX Triple Quad 6500+ system and the SCIEX 7500 system to compare sensitivity for analysis of SFTPs.
Quality control (QC) samples at low, mid and high concentrations were also assessed. QC low, mid and high were prepared at 8000 ng/mL, 800 ng/mL and 100 ng/mL, respectively. A total of 4 replicates were analyzed for each QC level.
ABFP provided an effective enrichment method for a multitude of signature peptides in human plasma. Separation analysis using microflow LC with trap-and-elute setup enabled increased sampling efficiency enabling sensitive analysis of critical protein biomarkers. Furthermore, coupling microflow LC analysis to the SCIEX 7500 system with front-end improvements such as increase in ion generation, capture and transmission enabled quantification of low-level targets at ng/mL levels.
Quantification results
ABFP employs a solid-phase extraction (SPE) cleanup along with an orthogonal fractionation step to separate the peptide targets. ABFP was compared with direct analysis to assess the level of improvement in sensitivity for hSFTP-C (Figure 2). ABFP achieved a 5-fold S/N improvement at 50 ng/mL over traditional direct analysis. This demonstrates that the SPE fractionation method substantially improves the sensitivity for quantification of signature peptides through targeted enrichment.
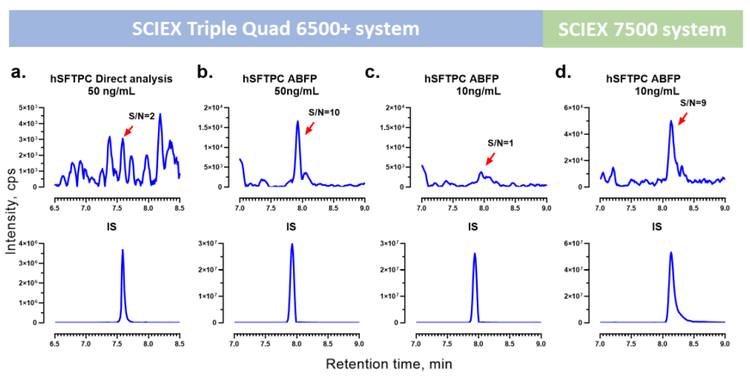
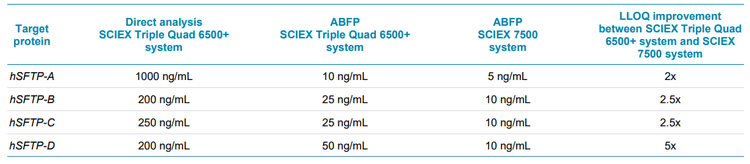
In addition, a comparative analysis was performed between the SCIEX 7500 system and the SCIEX Triple Quad 6500+ system using hSFTP-C at a concentration of 10 ng/mL. The application of the SCIEX 7500 system resulted in a S/N enhancement of up to 9-fold compared with the previous generation instrument (Figure 2). As a result, the LLOQs were improved by a factor of 2x to 5x for the hSFTPs using the SCIEX 7500 system (Table 5).
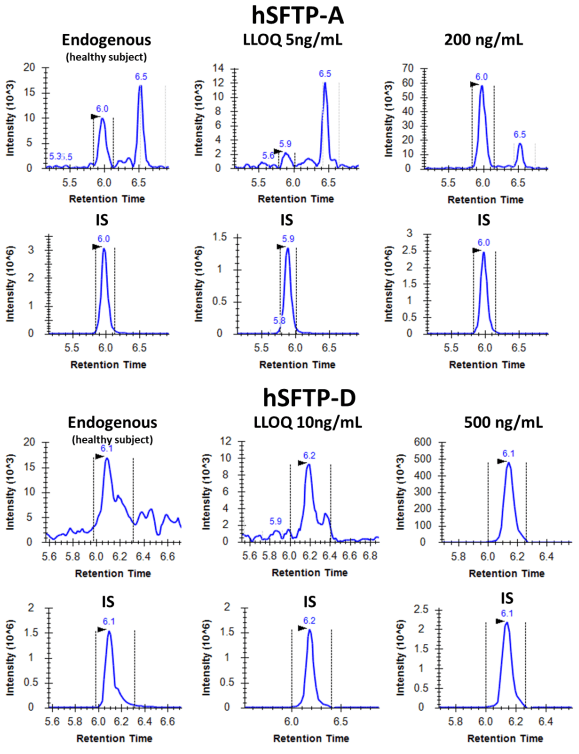
Figure 3 shows the representative XICs at the LLOQ levels of hSFTP-A and hSFTP-D. An LLOQ of 5 ng/mL and 10 ng/mL were achieved for hSFTP-A and hSFTP-D, respectively.
A highly sensitive, reproducible and efficient analysis of hSFTPs was demonstrated using ABFP enrichment coupled to a microflow LC and SCIEX 7500 system supporting quantification of low-level targets in early drug discovery and development.
Conclusion
- Low-ng/mL LLOQs were achieved for quantification of hSFTPs in human plasma using the SCIEX 7500 system with improved front-end technology for ion generation, capture and transmission
- A 2x to 5x improvement in LLOQ were reached using the SCIEX 7500 system compared with the SCIEX Triple Quad 6500+ system
- Excellent linearity, accuracy and %CV were achieved for the quantification of hSFTPs using the SCIEX 7500 system
References
- Bo An, Timothy W Sikorski, John F Kellie, Zhuo Chen, Nicole A Schneck, John Mehl, Huaping Tang, Jun Qu, Tujin Shi, Yuqian Gao, Jon M Jacobs, Eshani Nandita, Remco van Soest and Elliott Jones (2022). An antibody-free platform for multiplexed, sensitive quantification of protein biomarkers in complex biomatrices. Journal of Chromatography A, 1676: 463261.



