Abstract
This technical note demonstrates the application of a high-resolution mRNA analysis kit for the rapid separation of circular RNA (circRNA) species with a pre-assembled, bare-fused silica multi-capillary cartridge and a high-throughput electrophoresis system (BioPhase 8800 system). RNA co-analysis using a simplified sample preparation yielded high-resolution separation of circRNA species from linear RNA products. This approach also detected complex species, such as RNA concatemers, lowlevel impurities and fragmentation, by utilizing capillary gel electrophoresis with a laser-induced fluorescence detector (CGE-LIF).
Introduction
Circular RNA has gained increased attention in drug development and as a therapeutic target in recent years because its covalently closed loop structure provides exceptional stability and resistance to RNases.1,2 The production of circRNA-based therapeutics necessitates the implementation of an analytical method capable of separating the linear precursor and circRNA.3
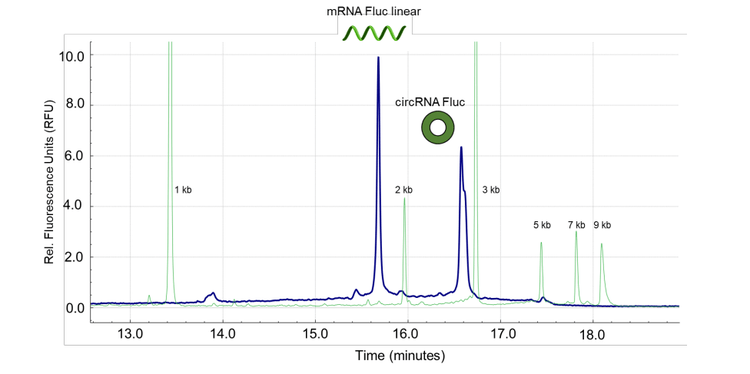
This technical note shows the separation of 2 synthesized circRNA products from their linear precursors using multi-capillary electrophoresis and the RNA 9000 Purity & Integrity kit.4 The circRNA species demonstrated delayed migration compared to the linear precursors (Figure 1). These results revealed the presence of putative RNA concatemers and provided information about the purity of circRNA species. The purity of the circRNA-Fluc product was 84.49% with a CV% value of 0.46% for 3 replicate analyses. Further studies using CGE-LIF detected linear mRNA-Fluc impurities spiked at levels as low as 0.1% of the final circRNA-Fluc product and yielded a linear dynamic range that spanned 3 orders of magnitude.
Key features of circRNA analysis by CGE-LIF
- circRNA products were differentiated from their linear precursors and other high molecular weight species with high resolution to test purity
- Precise and robust circRNA purity analysis with %CV values <2.0% for 3 replicate analyses
- Automated size determination for the linear precursor of the circRNA product
- Highly sensitive impurity detection with a linear dynamic range spanning 3 orders of magnitude
Methods
Materials: The RNA 9000 Purity & Integrity kit (P/N: C48231) containing the nucleic acid extended range gel, SYBRTM Green II RNA Gel Stain,* acid wash (regenerating solution), CE-grade water and the ssRNA Ladder (0.05-9 kb) was from SCIEX (Framingham, MA). The pre-assembled BioPhase BFS capillary cartridge (8 capillaries, 30 cm total length, P/N: 5080121), disposable BioPhase starter plate pack (P/N: 5080311) and the sample loading solution (SLS, P/N: 608082) were also from SCIEX.
Linear and circular RNA products: The linear and circular in vitro transcribed (IVT) RNAs were obtained from Creative Biolabs (Shirley, NY). Linear IVTScrip mRNA-Fluc (1.929 kb) and IVTScrip circRNA-Fluc encoded the luciferase gene (Fluc). The linear RNA-Fluc was synthesized with specific termini for ligation. Linear IVTScrip mRNA-eGFP (0.996 kb) and IVTScrip circRNA eGFP encoded enhanced green fluorescent protein (eGFP).
The linear Fluc and eGFP RNA products were certified by gel analysis, the capping assay and UV-VIS (OD 260/280) by the vendor. The Fluc and eGFP circRNA products were certified by the RNase R resistance test, ligation site and sequencing. The linear mRNA-Fluc and mRNA-eGFP products or precursors were shipped in a 1mM sodium citrate buffer, pH 6.4 at 1 mg/mL. The circRNA products for Fluc and eGFP were lyophilized at 10 μg and reconstituted at 1 μg/μL with nuclease-free water. The precursor and circular RNA products were stored at -80°C until the time of analysis.
Linear and circular RNA sample preparation
Integrity and purity analysis: To assess the integrity and purity of the linear Fluc and eGFP RNA products, working solutions of the linear products were separately prepared by diluting each product down to 9.77x10-2 ng/mL using a 1:1, water/SLS solution. The working solutions for the Fluc and eGFP circRNA products were prepared by diluting each product down to 3.91x10-1 ng/mL using a 1:1, water/SLS solution. Then, 50 μL aliquots of these working solutions were heated for 5 minutes at 70°C, immediately placed on ice and cooled for at least 10 minutes. Samples were then transferred to the multi-capillary 96- well sample plate and analyzed using the BioPhase 8800 system and the RNA 9000 Purity & Integrity kit (Figure 2) with the separation method illustrated in Figure 3.
Separation of circRNA from the linear precursor: For the co-analysis of the linear and circular RNA-Fluc products, a working stock solution of the linear Fluc product was prepared at 1.56 ng/mL by serial dilution of the main stock (1 mg/mL). The working stock solution for the circRNA-Fluc product was prepared by serial dilution of the main stock (1 mg/mL) to a final concentration of 6.25 ng/mL. Next, ~3.13 μL of the linear and of the circular RNA working solutions were added to 43.75 μL of 1:1, water/SLS to yield a 50 μL solution containing 9.77x10-2 ng/mL of the linear RNA and 3.91x10-1 ng/mL of the circRNA. Samples were heated for 5 minutes at 70°C, immediately placed on ice and cooled for at least 10 minutes. Next, samples were transferred to the 96-well sample plate and analyzed using the multi-capillary BioPhase 8800 system and the RNA 9000 Purity & Integrity kit.
The linear and circular RNA-eGFP products were mixed as described for the co-analysis of the linear and circular RNA-Fluc products. The final concentrations of the linear and circular RNAeGFP products were 9.77x10-2 ng/mL and 3.91x10-1 ng/mL, respectively. The separation parameters, conditioning method and shutdown procedures outlined in Figure 3 were used with a 30-min runtime.
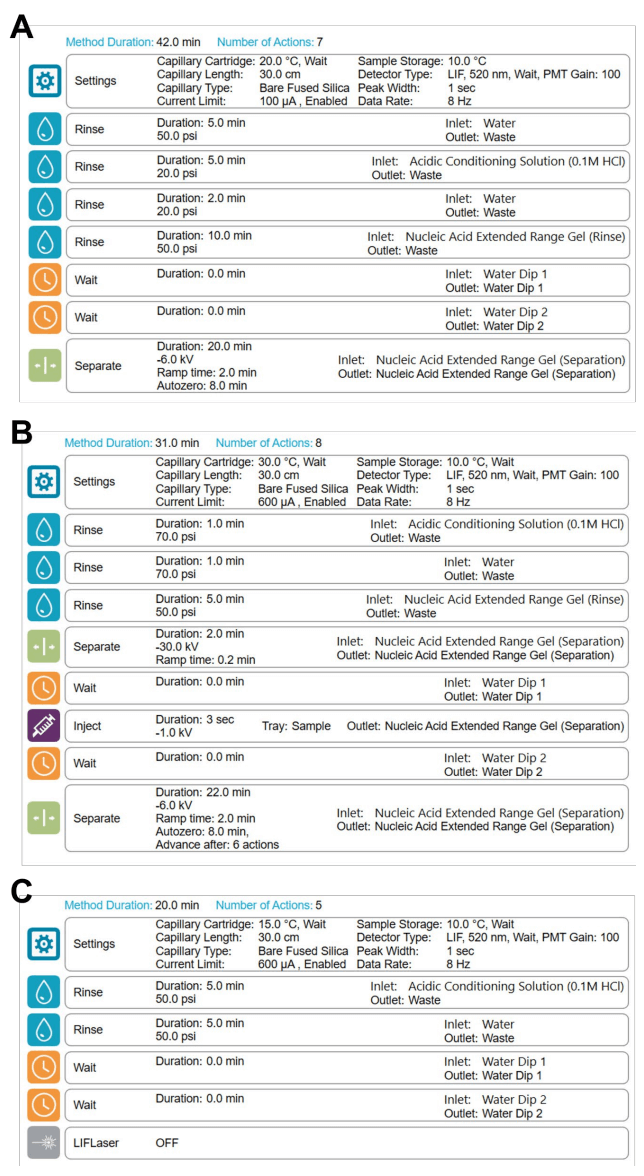
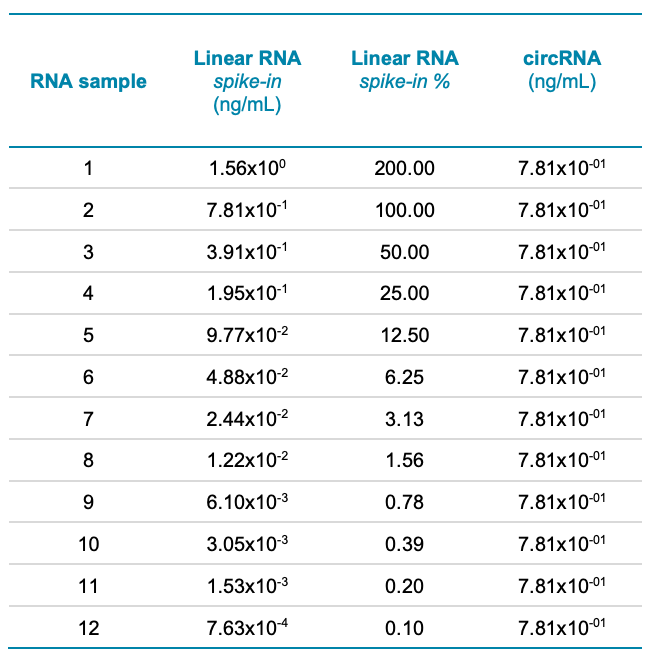
Spiking sensitivity assay: In this experiment, linear RNA-Fluc (precursor) was spiked into circRNA-Fluc to simulate incomplete circularization to circRNA-Fluc. The amount of circRNA-Fluc was kept constant at 7.81x10-1 ng/mL for this assay and linear RNAFluc was spiked in as an impurity at concentrations ranging from 7.63x10-4 mg/mL to 1.56 ng/mL. Linear RNA-Fluc was serially diluted 2-fold to yield 12 concentrations. Sensitivity was assessed at the 0.10% impurity level and the linear dynamic range was calculated (Table 1). All working solutions and mixtures were prepared using 1:1, water/SLS. Samples were heated for 5 minutes at 70°C, immediately placed on ice and cooled for at least 10 minutes before transferring the samples to the multi-capillary 96-well sample plate for analysis with the BioPhase 8800 system and the RNA 9000 Purity & Integrity kit.
Working solutions were prepared and sample preparation steps were performed on ice to minimize sample degradation. Freezing/thawing cycles were avoided by making aliquots.
RNA ladder sample preparation: The RNA ladder sample was prepared as described in the user manual for the RNA 9000 Purity & Integrity kit.4 Briefly, 2 µL of the ladder was mixed with 48 µL of SLS and then heated for 5 minutes at 70°C using a thermal cycler. Samples were then chilled on ice for at least 5 minutes and 60 µL of CE-grade water was added before transferring to the multi-capillary sample plate.
Instrument and software: A BioPhase 8800 system (P/N: 5083590) equipped with a LIF detector was from SCIEX. The excitation and emission wavelengths used were 488 nm and 520 nm, respectively. Data acquisition and analysis were performed using the BioPhase 8800 software v1.2.20 e-license (SCIEX).
Results
Integrity and purity analysis for linear RNA-Fluc
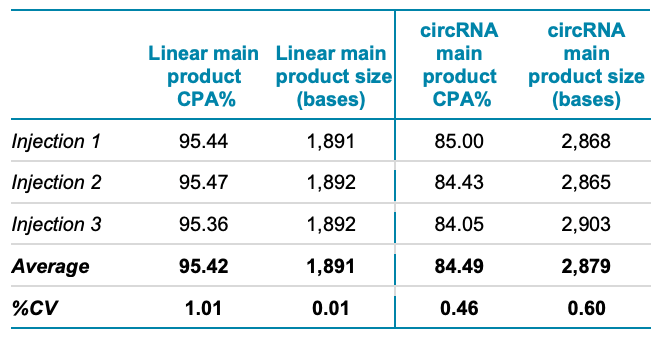
The termini for ligation on the linear RNA-Fluc were examined for integrity and purity using the BioPhase 8800 system and RNA 9000 Purity & Integrity kit. Figure 4A shows the overlay of 3 sequential injections of the linear mRNA-Fluc with an expected size of 1.929 kb. The linear mRNA-Fluc (pink traces) mainly showed 2 products, labeled A and B in the electropherogram. Peak B is the main RNA-Fluc peak that migrates before the 2 kb marker, whereas peak A might correspond to an impurity in the RNA product.
The product composition of the linear mRNA-Fluc was then tested using the BioPhase 8800 system. Table 2 shows the purity of the linear mRNA-Fluc product based on the CPA% of triplicate injections (Figure 4A). The linear mRNA-Fluc had an average purity of 95.42% with a %CV of 0.05% (Table 2).
The BioPhase 8800 system was used to determine the size of the main observed peak (peak B) of linear mRNA-Fluc to confirm its identity. The point-to-point fit type using 10 RNA size markers from the RNA 9000 ladder indicated a main product peak size of 1.891 kb (Table 2).
Integrity and purity analysis for circRNA-Fluc
A circRNA product was analyzed by CGE-LIF to examine its purity. The circRNA-Fluc product contained more impurities and nucleic acid fragments than the linear mRNA-Fluc product, indicating the presence of circRNA. HMW products were also present, which might correspond to RNA concatemers.
Figure 4B shows the overlay of 3 sequential injections (orange traces) of circRNA-Fluc after enzymatic ligation. The peaks corresponding to the main products are labeled C and D. Impurities detected included a HMW product (peak E) and 2 low molecular weight products (peaks A and B). The presence of these impurities indicates that the circularization reaction might have resulted in multiple impurities and topologies of the circRNA. Automated CPA% calculations indicated that the purity of the circRNA-Fluc product was 84.49% (n=3, %CV of 0.46%).
The results shown in Figure 1 illustrate the separation of the linear (9.77x10-2 ng/mL) and the circular (3.91x10-1 ng/mL) RNA-Fluc products (blue trace) in a co-analysis test. Consistent with the individual characterization of the linear precursor (Figure 4A) and the circRNA (Figure 4B), the co-analysis of these 2 molecules demonstrated the successful separation of the linear precursor and the circRNA products. In summary, this co-analysis supports a fast and efficient method to assess circRNA products, impurities, HMW species and putative RNA concatemers.
Separation of linear and circular RNA-eGFP
This workflow was tested on linear and circular RNA-eGFP to confirm its ability to separate small linear and circular RNA products.
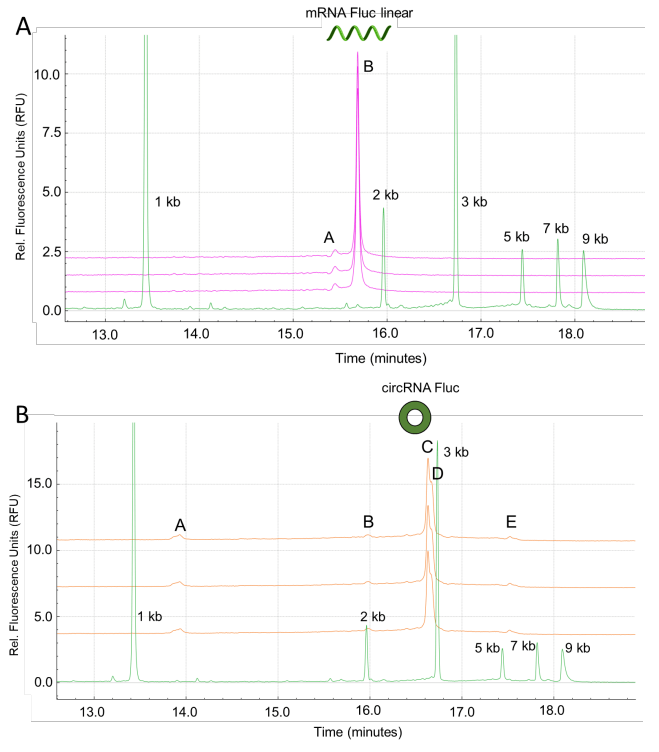
Figure 5A illustrates the purity profiling of the linear RNA-eGFP product (pink traces). The overlay of 3 injections confirmed the purity analysis and migration pattern for the linear RNA product with a determined size of 1,190 kb using the BioPhase Analysis tool.
Figure 5B reports the purity profiling of the circRNA-eGFP product with 3 HMW species (peaks C, D and E). The percent composition based on CPA% from 3 measurements for these fragments was 4.78% (%CV of 7.28%) for peak A, 21.64% (%CV of 1.13%) for peak B, 1.80% (%CV of 4.71%) for peak C, 1.24% (%CV of 34.15%) for peak D, 70.30% (%CV of 0.24%) for peak E and 0.23% for impurities migrating before product A.
The electropherogram results and BioPhase 8800 system analysis of this injection confirmed the migration patterns and main product composition observed in the analysis of individual linear and circular RNA-eGFP products (Figure 5C). These results demonstrate the simultaneous evaluation of linear and circular RNA-eGFP at 9.77x10-2 ng/mL and 3.91x10-1 ng/mL, respectively. The size for the linear RNA-eGFP was 1.190 kb (n=3, %CV of 0.34%).
circRNA-Fluc 0.1% sensitivity analysis
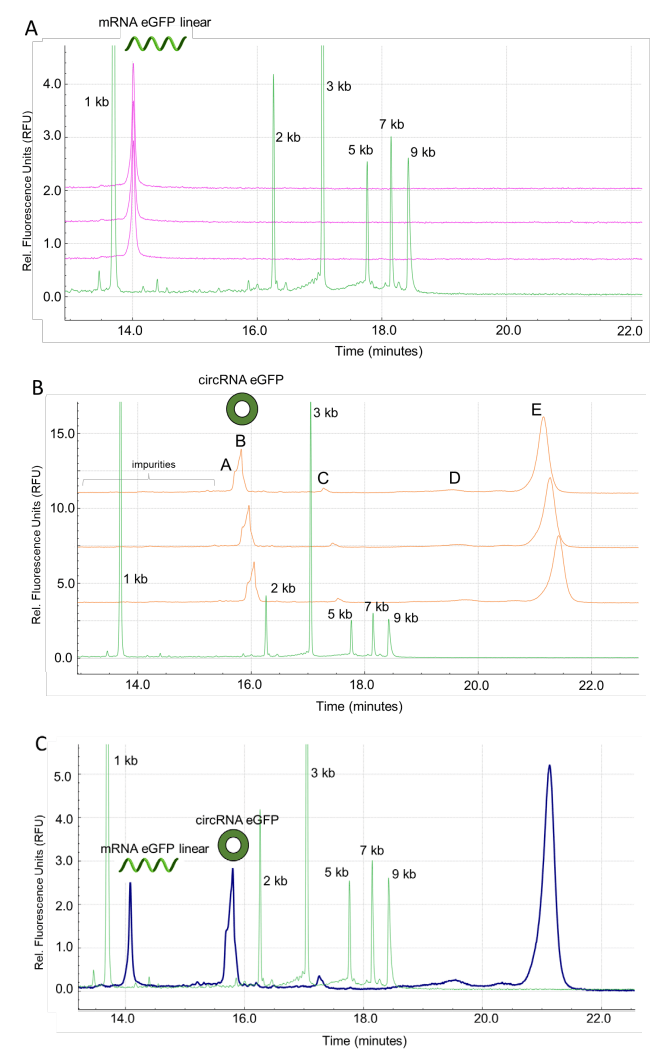
The sensitivity of the workflow was tested by spiking linear RNA-Fluc into a set concentration of circRNA-Fluc. Figure 6A shows representative electropherograms at varying linear RNA-Fluc spike concentrations. Figure 6B shows a zoomed-in view to highlight the detection of linear Fluc-RNA at impurity levels <1%. In summary, this analysis demonstrated the capability of the BioPhase 8800 system to achieve the detection of the linear RNA-Fluc product at concentrations ranging 3 orders of magnitude, from 7.63x10-4 ng/mL to 1.56 ng/mL.
Conclusion
- Separation of circRNA from the linear precursor was demonstrated for Fluc and eGFP mRNAs using a simple workflow
- Unexpected HMW RNA species in the circRNA final products were identified by automated size and purity composition analysis
- Detection of impurities present at 0.1% of the main product was demonstrated in this study
References
- Liu, X., Zhang, Y., Zhou, S., Dain, L., Mei, L., and Zhu, G. Circular RNA: an emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. Journal of Controlled Release. 2022, 348, 84-94
- Wesselhoeft, R. A., Piotr, S.K., Anderson, G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nature Communications. 2018. 9. 2629. 1-10.
- Abe, B.T., Wesselhoeft, R. A., Chen, R., Anderson, D., and Chang, Y. H. Circular RNA migration in agarose gel electrophoresis. Molecular Cell, Matters Arising. 2022. 82. 1768-1777.
- RNA 9000 Purity & Integrity Kit for the BioPhase 8800system Application Guide