Genome integrity analysis of adeno-associated viruses (AAV) using multi-capillary gel electrophoresis
Tingting Li, Jane Luo, and Sahana Mollah
SCIEX, USA
Introduction
Rapid and robust analysis of adeno-associated virus (AAV) genome integrity was achieved by simultaneous separation of multiple samples on a multi-capillary analyzer, the BioPhase 8800 system. This increased capacity enables parallel processing of AAV genome integrity analysis with excellent sensitivity and repeatability.
An AAV is a non-enveloped virus with an icosahedral protein shell capsid of approximately 20 nm in diameter and a singlestranded DNA genome of about 4.7 kb in length. Due to its lack of pathogenicity, low immunogenicity, broad tropism and persistent transgene expression in both proliferating and quiescent cells, AAV is an attractive choice for creating viral vectors for gene therapy for a variety of diseases. The genome of an AAV vectorfor gene therapy is usually composed of two inverted terminal repeats (ITR), a promoter, a transgene and a poly-A tail. AAV genome integrity analysis is a critical quality test for AAVs because it provides insights into transgene integrity and ensures product safety and efficacy.1
Verification of genome size has been traditionally done by denaturing agarose-gel electrophoresis and southern blot.2 Both are time consuming and have limited resolution on size determination.2,3 Recently, a capillary electrophoresis with laser induced fluorescent (CE-LIF) detection method was developed for AAV genome integrity.4 CE-LIF is a well-established, high sensitivity technique for nucleic acid analysis. Currently, AAV genome integrity analysis by CE-LIF is performed one sample at a time using the single-capillary system. Multiplexing the analysis can help decrease the analysis or profiling time for multiple serotypes or AAV vectors with different genome sizes.
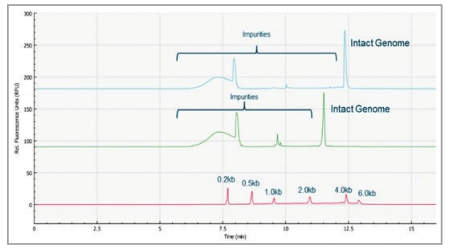
Figure 1. Genome size analysis of AAV2 samples of different genome loads on the BioPhase 8800 system. Red trace: RNA size standards with sizes marked in dark blue font. Green trace: AAV2-CMV-GFP sample with genome size about 2.4 kb. Blue trace: AAV2-CMV-Lacz sample with genome size about 4.7 kb.
Figure 2. The BioPhase 8800 system equipped with LIF and UV detectors and consumable/reagent kits.
In this technical note, it is demonstrated that multiple AAV samples with different serotypes or different genome sizes can be run simultaneously on the multi-capillary BioPhase 8800 system to accelerate the execution of sensitive, AAV genome integrity analysis while retaining excellent resolution, sensitivity and repeatability as seen with the single-capillary PA 800 Plus.
Key features
- Easy transfer of methods from the single-capillary PA 800 Plus to the multi-capillary BioPhase 8800 system
- Quick assessment of AAV genome size purity with multiplex screening capability
- Excellent separation resolution of intact from partial AAV genomes and small size impurities
- Good repeatability with %RSD of migration time and % corrected area of <1% and <5%, respectively
- Capability of analyzing the genome integrity of AAV samples of multiple serotypes or different genome sizes in one analysis
Materials and methods
Materials
Chemicals: Urea (P/N 29700), SYBR Green II RNA Gel Stain, 10,000X concentrate in DMSO (P/N S7564), and nuclease-free water (P/N AM9932) were obtained from Thermo Fisher Scientific,Waltham, MA. Polyvinylpyrrolidone (P/N 437190) and TBE Buffer,10X, Molecular Biology Grade (P/N 574795) were obtained from Sigma- Aldrich, St Louis, MO. QIA quick PCR purification kit (P/N 28104) was from Qiagen, Germantown, MD. Sample Loading Solution (P/N 608082) was from SCIEX, Framingham, MA.
Samples: Packaged AAV8 of pAV-CMV-BuB1, Packaged AAV8 of pAV-CMV-BuB1 Empty Capsid, Packaged AAV8 of pAV-CMVGFP and Packaged AAV8 of pAV-CMV-GFP Empty Capsid were from Vigene Biosciences, Rockville, MD.
AAV2-CMV-GFP (P/N SL100812), AAV2-CMV-Lacz (P/N SL100854), AAV5-CAG-GFP (P/N SL100823), AAV5-CMV-Lacz (P/N SL100857), AAV8-CAG-GFP (P/N SL100837), AAV9-CMVNull (P/N SL100839), AAV9-CAG-GFP (P/N SL100844) and AAV9-CMV-Lacz (P/N SL100868) were from Signagen, Rockville, MD.
RNA 6000 Ladder (P/N AM7152) was obtained from Thermo Fisher Scientific, Waltham, MA.
Buffer preparation
Preparation of 1x TBE rinse buffer. 10 mL of 10X TBE buffer was added into 90 mL of nuclease-free water followed by vortexing.
Preparation of 100 times diluted Sybr Green II. 10 µL of Sybr Green II was diluted in DMSO.
Preparation of the PVP gel separation buffer. The separation buffer was 1% PVP (1.3 MDa) in 1x TBE buffer (89 mM Tris, 89 mM boric acid, 2mM EDTA, pH 8.3) with 4 M urea and 0.2% SYBR green dye. 24.024 gram of urea, 10 mL of 10x TBE buffer were then added to 50 mL of nuclease-free water. The solution was stirred for 1-2 hours at room temperature until the solid was totally dissolved. It was then transferred into a 100 mL volumetric flask and filled to the mark with a sufficient quantity of nucleasefree water. The solution was then transferred into an amber bottle containing 1.000 gram of polyvinylpyrrolidone (PVP). The buffer was slowly stirred at 4°C overnight to fully dissolve the PVP polymer,and was then filtered using a 0.45 μm filter. This buffer was stored in a refrigerator (2°C - 8°C) in 20 mL aliquots. Before the sample run, the aliquot of buffer was mixed with 80 μL of 100x diluted Sybr Green II solution on the day of usage.4
Sample preparation
RNA 6000 Ladder. For the reproducibility study, 50 μL of the RNA 6000 ladder was diluted in 350 μL of nuclease-free water and mixed with 400 μL of sample loading solution, heated at 70°C for 10 minutes and then immediately placed on ice.5 Diluted ladders of RNA 6000 were subjected to a similar denaturation procedure before being loaded onto the instrument as RNA size markers.
AAV samples. The AAV samples were prepared following a simplified workflow for genome release and purification. 20 μL of the each AAV sample was mixed thoroughly with the binding buffer from the QIAquick PCR purification kit. The manufacturer’s instructions in the kit were followed except for the step where the column was washed twice. The AAV genome sample was eluted from the column using 50 μL of 10x diluted by elution buffer (Buffer EB) from the kit. Before loading onto the instrument for analysis, 10 μL of the eluted AAV genome solution was mixed with 40 μL of nuclease-free water and 50 μL of sample loading solution, heated at 70°C for 10 minutes and immediately cooled on ice for 5 minutes.
For AAV samples with residual host cell nucleic acids, a longer sample preparation workflow was used. The protocol includes benzonase treatment to degrade small sized impurities outside of AAV capsids, filtration to remove the benzonase and degraded nucleic acids and proteinase K treatment for AAV genome release from the capsids prior to the QIAquick PCT purification.4
The simplified workflow was used for the AAV sample set used in this technical note since the purchased AAV samples have already been treated with benzonase for host cell nucleic acids removal during the product purification process by the vendors.
Instrument setup
All single-capillary electrophoresis analyses were performed on the PA 800 Plus Pharmaceutical Analysis System. EZ cartridge re-assembled with bare fused-silica capillary (50 μm I.D., 30 cm total length, 20 cm effective length) was purchased from SCIEX (PN A55625, Framingham, MA). Samples were introduced into the inlet of the capillary electrokinetically at -5 kV for 10s. Separations were performed using reversed polarity with 300 V/cm electrical field at 25° C. Sample trays were kept at 4°Cto minimize RNA degradation and renaturation. LIF detector was configured with a 488-nm laser with an emission filter of 520nm.
The multi-capillary separations utilized the BioPhase 8800 system equipped with LIF detector (with 520 nm emission filter). The separation parameters and methods were the same as those in the single-capillary analyses and are indicated in Figures 3, 4, and 5. The separations were accomplished in the 8 channels, 30 cm total length each, BioPhase BFS Capillary Cartridge (Part # 5080121, SCIEX). BioPhase Analysis software was then used for data processing.
Figure 3. Cartridge conditioning method as shown on theBioPhase 8800 system.
Figure 4. Separation method as shown on the BioPhase 8800 system.
Figure 5. Shutdown method as shown on the BioPhase 8800 system.
Results and discussion
Comparison between the BioPhase 8800 system and the PA 800 Plus
To compare the data acquired between the multi-capillary and the single-capillary platforms, analysis of 3 different AAV serotypes (serotype 2, 5, and 9) was performed on both the PA 800 Plus and BioPhase 8800 systems. The genome profile and the migration times of the nucleic acid peaks align well between the two systems as shown in Figure 6. Additionally, as shown in Table 1, the % corrected peak areas of the intact genome and the impurities (including truncated genome and other small sized nucleic acid impurities) on the two systems correlate well with each other.
Figure 6. Comparison of profiles from genome integrity analysis of AAV2, AAV5 and AAV9 on the single-capillary PA 800 Plus and the multicapillary BioPhase 8800 system using a LIF detector. Results obtained on both instruments correlate well.
Table 1. Comparison of Corrected Peak Area% of AAV2, AAV5, and AAV9 on the single-capillary PA 800 Plus and the multi-capillary BioPhase 8800 system. Results obtained on both instruments correlate well with each other.
Reproducibility of migration time and corrected peak area%
Figure 7 shows 8 electropherograms of RNA ladder analysis, simultaneously acquired on the 8 capillaries of the multi-capillary electrophoresis system. High-resolution separation of all RNA size markers (0.2 kb, 0.5 kb, 1.0 kb, 2.0 kb, 4.0 kb, and 6.0 kb) were obtained. The reproducibility of 10 consecutive injections of the RNA ladder sample on 8 capillaries was evaluated as well. Figure 8 shows the overlaid 80 electropherogram traces from 10 consecutive injections of the RNA ladder sample on all 8 capillaries. The migration time reproducibility (RSD%) of the 80 analysis for each RNA size marker was less than 1%, as depicted in Table 2. The corrected peak area% reproducibility (RSD%) of 80 injections was <5% for the RNA markers.
Figure 7. Multi-capillary electrophoresis of RNA size ladder. Traces A to H represent the separation in the eight individual capillaries of the BioPhase 8800 system. RNA size markers of 0.2 kb, 0.5 kb, 1.0 kb, 2.0 kb, 4.0 kb, and 6.0 kb are well separated.
Figure 8. Overlaid traces of 80 injections (10 consecutive injections of 8 capillary channels) of RNA size ladder on the multi-capillary electrophoresis system.
Table 2. Reproducibility (RSD%) of migration time (MT) and corrected area% (CA%) of 80 injections of each RNA maker in the RNA size ladder on the BioPhase 8800 system.
Analysis of AAV vectors of different serotypes
The AAV samples of different serotypes can be loaded onto different capillary channels and analyzed simultaneously on the BioPhase 8800 system. As shown in Figure 9 AAV samples of different serotypes (Serotype 2, 5, 8, and 9) were analyzed using the multi-capillary electrophoresis system. The red trace is the RNA size standards with sizes marked in dark blue font. The green trace is the AAV2-CMV-Lacz sample. Light blue trace is the AAV5- CMV-Lacz sample. The dark blue trace is AAV8- CMV-BuB1 sample. And the pink trace is AAV9-CMV-Lacz sample. The intact genome of AAV was well separated from the partial or truncated genome and other small size impurities for different serotypes of AAV samples. Compared to multiple hours of the analysis time of analyzing a number of AAV samples on the single-capillary electrophoresis system, it takes less than 25 minutes for screening of 8 samples on the BioPhase 8800 system.
Figure 9. Genome Integrity Analysis of AAV samples of different serotypes (Serotype 2, 5, 8 and 9) done in parallel on the BioPhase 8800 system. Red trace:RNA size standards with sizes marked in dark blue font. Green trace: AAV2-CMV-Lacz sample. Light blue trace: AAV5- CMV-Lacz sample. Dark blue trace: AAV8-CMV-BuB1. Pink trace: AAV9-CMV-Lacz sample.
Analysis of AAV vectors encapsulating different genome size
Figure 1 shows the analysis of AAV 2 samples encapsulating different genome sizes. The red trace is RNA size standards with sizes marked in dark blue font. The green trace is the electropherogram of AAV2-CMV-GFP sample with genome size of~2.4 kb. The blue trace is the electropherogram of AAV2- CMV-Lacz sample with genome size of about 4.7 kb. The genome size difference of these two AAV 2 vectors was observed in the electropherograms. It is worth noting that the RNA size standards migrate slower in this PVP gel buffer than the single stranded AAV genome of the same size due to the differences in base composition in these nucleic acids, and the differences related to ribose in RNA versus deoxyribose in single stranded DNA.
Analysis of AAV full and empty capsid samples
The enriched AAV full capsids and enriched AAV empty capsids of AAV8-CMV-BuB1 were analyzed using this PVP gel-based CGE method on the BioPhase 8800 system as shown in Figure 10. The red trace is the RNA size standards with sizes marked in dark blue font. The blue trace is the enriched full capsid AAV8-CMV-BuB1 sample. The green trace is the enriched empty capsid AAV8-CMV-BuB1 sample. A small amount of intact genome was observed in the enriched empty AAV8- CMV-BuB1 sample which indicated the present of small amount full capsids in the enriched empty capsids sample.
Figure 10. Genome Integrity Analysis of enriched full capsids and enrichedempty capsids of AAV8-CMV-BuB1 on the BioPhase 8800 system. Red trace: RNA size standards with sizes marked in dark blue font. Blue trace: Enriched full capsids of AAV8-CMV-BuB1 sample. Green trace: Enriched empty capsids of AAV8-CMV-BuB1 sample.
Conclusions
- In this technical note, the BioPhase 8800 system demonstrates excellent resolution and comparable %purity of AAV genome size analysis to the PA 800 Plus. This makesan easy and seamless transition of this CGE-LIF method for genome integrity analysis from a single-capillary to amulti-capillary platform
- The multiplexing capability of the BioPhase 8800 system enablesfaster assessment of genome purity of multiple AAV samples by accelerating screening or process development of AAV products
- Capability of quick assessment of genome integrity of AAV samples of different serotypes or different genome payloads,with 15 minutes sample preparation and 25 minutes analysis of 8 samples in parallel
- Good inter-capillary and intra-capillary reproducibility with %RSD of migration time and % corrected area of <1% and <5%, respectively
References
- Adeno-Associated Virus (AAV) as a Vector for Gene Therapy.(2017) BioDrugs 31:317–334.
- Characterization of Genome Integrity for Oversized Recombinant AAV Vector. (2010) Molecular Therapy 18(1):87–92.
- Viral Quantitative Capillary Electrophoresis for Counting IntactViruses. (2011) Analytical Chemistry 83(13):5431-5.
- Genome Sizing Methodology for Transgene Integrity Analysisof Adeno-Associated Viruses (AAV). SCIEX technical note, RUO-MKT-02-10925-A.
- Method optimization and evaluation for RNA purity analysisusing CE-LIF technology. SCIEX technical note, RUO-MKT-02-8017-B.
 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge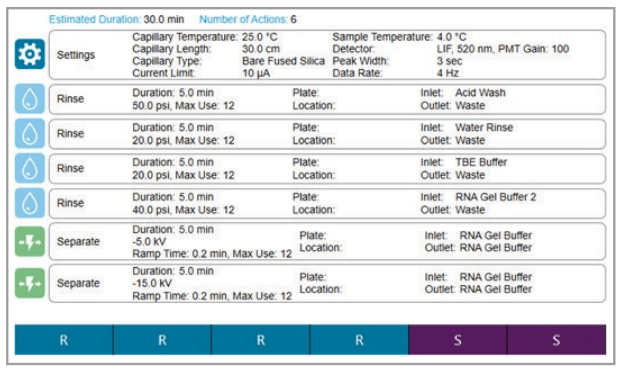 Click to enlarge
Click to enlarge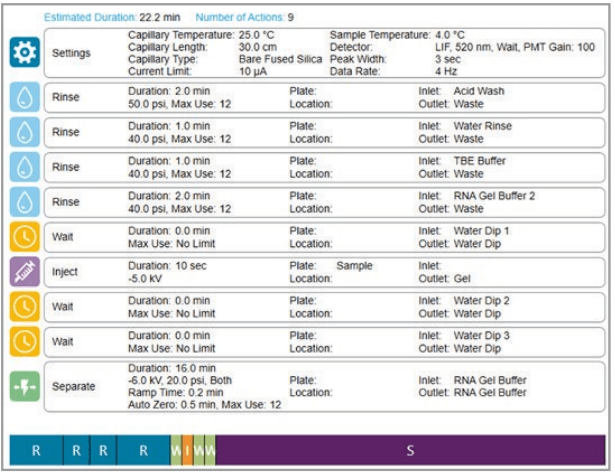 Click to enlarge
Click to enlarge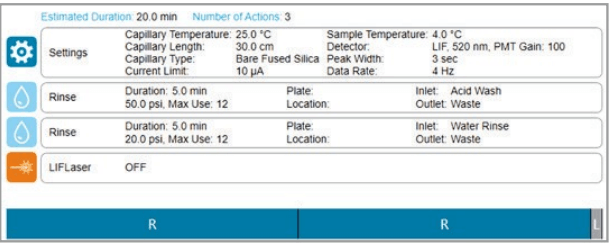 Click to enlarge
Click to enlarge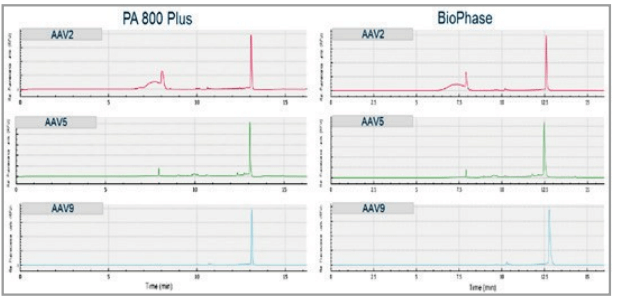 Click to enlarge
Click to enlarge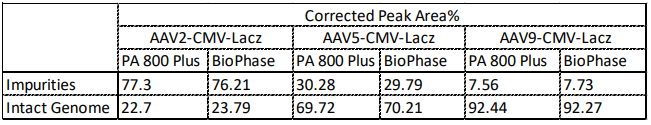 Click to enlarge
Click to enlarge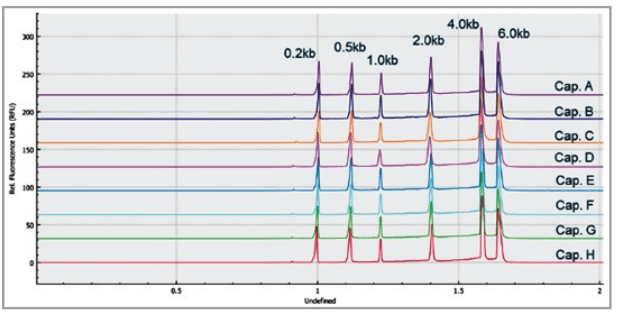 Click to enlarge
Click to enlarge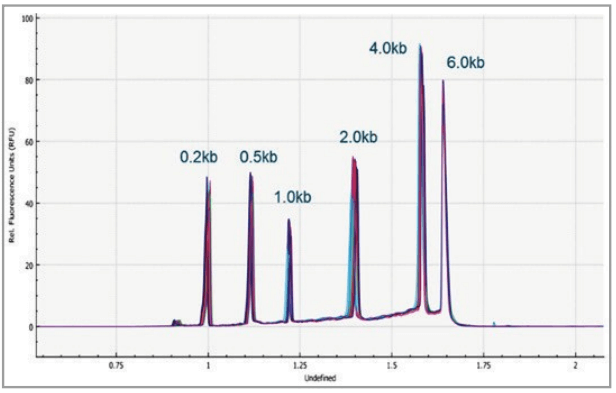 Click to enlarge
Click to enlarge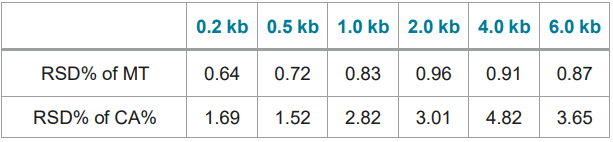 Click to enlarge
Click to enlarge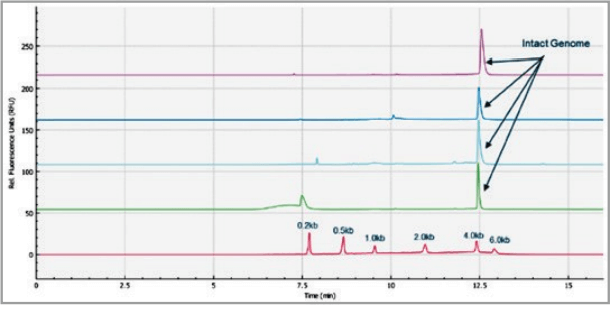 Click to enlarge
Click to enlarge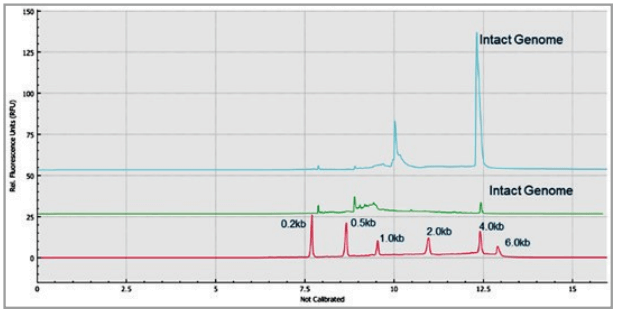 Click to enlarge
Click to enlarge