Genome sizing methodology for genome integrity analysis of adeno-associated viruses (AAV)
Jane Luo, Tingting Li, Fang Wang, Sahana Mollah, Handy Yowanto
SCIEX, Brea, California
Abstract
One of the most commonly used methods for the production of rAAV vectors is the triple-transfection method which involves co-transfection of permissive cells such as HEK293 cells with three plasmids: one containing the transgene of interest flanked by the AAV inverted terminal repeats (ITRs), a packaging plasmid containing rep and cap genes, and a third plasmid encoding adenoviral helper genes. Gradient ultra-centrifugation is often used for purification of packaged rAAV vectors (Full capsids) from cellular debris contaminants, host cell DNA and RNA as well as Empty AAV capsids. However, remnants of contaminants can still be present in the viral vector product after purification. In addition, errors made in genome encapsidation in rAAV production can lead to heterogeneous populations with both intact genome and partial or smaller than unit-length genomes. The quality of Full capsids has a direct impact on the efficacy of the treatment including outcome of both preclinical and clinical studies. Therefore, it is crucial to accurately assess the quality and the correct length/size of the genome encapsulated in the vector. In this technical note, we describe the development of novel CE-LIF based methods for accurate determination of size purity of AAV genome. Our results indicate that these methods are capable of detecting intact and partial AAV genome as well as small size impurities.

Introduction
Adeno-associated virus (AAV) is a small (25-nm) virus that is composed of a non-enveloped icosahedral protein shell called capsid and a single-stranded DNA genome of about 4.7 kb. With its excellent safety profile and high efficiency in transducing a broad range of target tissues, AAV vector has become an attractive choice for gene therapy.1 Two recombinant AAV (rAAV) based drugs have been approved by the FDA: Zolgensma by AveXis for spinal muscular atrophy and Luxturna by Spark Therapeutics for inherited blindness. Many more are in clinical trial. One of the most commonly used methods for the production of rAAV vectors is the triple-transfection method which involves cotransfection of permissive cells such as HEK293 cells with three plasmids: one containing the transgene of interest flanked by the AAV inverted terminal repeats (ITRs), a packaging plasmid containing rep and cap genes, and a third plasmid encoding adenoviral helper genes. Gradient ultra-centrifugation is often used for purification of packaged rAAV vectors (Full capsids) from cellular debris contaminants, host cell DNA and RNA as well as Empty AAV capsids. However, remnants of contaminants can still be present in the viral vector product after purification. In addition, errors made in encapsidation of the intact genome containing the transgene during rAAV production can lead to heterogeneous populations with both intact genome and partial or smaller than unit-length genomes. The quality of Full capsids has a direct impact on the efficacy of the treatment including outcome of both preclinical and clinical studies. Therefore, it is crucial to accurately assess the quality and the correct length/size of the genome encapsidated in the vector. Verification of genome size has been traditionally done by denaturing agarose-gel electrophoresis and southern blot.2 Both are time consuming and with limited resolution on size determination. In this technical note, we describe the development of novel CE-LIF based methods for accurate determination of size purity of AAV genome. Our results indicate that these methods are capable of detecting intact and partial AAV genome as well as small size impurities.
Figure 2. Genome Size Analysis of an AAV8 Sample. Blue trace: RNA size standards with sizes in kilobases marked in black numbers; Red trace: AAV8 sample; Green trace: blank control.
Key Features
- Excellent separation resolution between intact and partial AAV genomes from small size impurities
- Sample separation is complete within 15 minutes, compared to hours to days with denaturing agarose gel and multiple hours with southern blot
- Quick assessment of AAV genome size purity can be achieved in 30 minutes through a simplified, screening workflow, compared to hours with denaturing agarose gel or southern blot
- A more comprehensive workflow that provides a more definitive answer • Both the screening and comprehensive workflows are suitable for multiple AAV serotypes
- Excellent separation repeatability with CVs of less than 0.5% for migration time
Methods
Materials: The LIF Performance Test Mix (PN: 726022), Nano vials (PN 5043467) and pre-assembled EZ-CE Capillary Cartridge (PN A55625, Figure 1B) were from SCIEX, Framingham, MA. Urea (PN 29700), Nuclease-free water (PN AM9932), SYBR Green II RNA gel stain, 10,000x concentrate in DMSO (PN S7564), 10x DNase I buffer (PN AM8170G) were obtained from Thermo Fisher Scientific, Waltham, MA. Polyvinylpyrrolidone (PVP, PN 437190), benzonase (PN E1014-5KU), 0.5 M EDTA, pH 8.0 (PN E7889-100ML), Transcript RNA markers 0.2-10 kb (PN R7020) and 10x Tris Borate EDTA (TBE) buffer (PN 574795), Molecular Biology Grade, Amicon Ultra-0.5 centrifugal filter unit with MW cut off of 100 kDa (PN UFC510024) were from Millipore Sigma, St. Louis, MO. The 5 µm syringe filter (PN 4650) was from PALL Corporation, Port Washington, NY. Rainin LTS filter tips were from Mettler Toledo, Oakland, CA. QIAquick PCR purification Kit (PN 28104) and Proteinase K (PN 19131) were from Qiagen, Germantown, MD. Packaged AAV8 and AAV formulation buffer (1X PBS with 0.001% Pluronic F68) were from Vigene Biosciences, Rockville, MD. AAV5 and AAV2 were from Signagen, Rockville, MD.
Instrument and Software: A PA 800 Plus Pharmaceutical Analysis System (Figure 1A) equipped with LIF detector and solid-state laser with excitation wavelength at 488 nm and a 520 nm band pass emission filter were from SCIEX, Framingham, MA. Data acquisition and analysis were performed using 32 Karat™ Software 10.3.
LIF Calibration: To ensure consistent response of LIF detector throughout this study, the LIF detector was calibrated using LIF Calibration Wizard and Performance Test Mix (PN 726022) following the instructions in the PA 800 Plus System Maintenance Guide (PN A51964).
Sample Storage: AAV samples and the Sigma Transcript RNA Markers were aliquoted at 5 to 20 µL upon first thawing and stored at -80oC freezer to avoid multiple freeze-thaw cycles.
Sample Preparation for RNA Markers: The Sigma Transcript RNA markers were diluted in nuclease-free water to 1 ng/µL, heat-treated and loaded onto the instrument as described above for the AAV genome sample.
Preparation of Buffer Trays and Sample Trays: All solutions were pipetted with filter tips. Each “NF Water” vial was filled with 1.5 mL nuclease-free (NF) water. Waste vial was filled with 1 mL NF water. “Sep Buffer” and “TBE Buffer” vials were filled with 1.5 mL of separation buffer containing SYBR Green II dye or 1x TBE.
Results and Discussions
Analysis of AAV Genome of Different Sizes: Packaged AAV8 of pAV-CMV-GFP (Vigene) and AAV5 of pAAV-CMV-GFP (Signagen) were treated with proteinase K to release the AAV genome.3 Nucleic acids purified with QIAquick from these samples were analyzed by CE-LIF on a PA 800 Plus Pharmaceutical Analysis System with a sample injection condition of 5 kV for 3 seconds. In Figure 3, the red trace was obtained with the RNA size standard. The blue trace was obtained with the AAV8 genome sample at 5 µL with a titer of 1.10 x 1013 GC/mL. The intact genome (2.8 kb) as well as the small size impurities between 0.5 to 1 kb were also detected. In addition, a small peak slightly larger than 2.8 kb was present. This may be the same intact genome but with secondary structure. There is also a shallow peak larger than 10 kb. Further experiments need to be conducted to determine if this was due to a multimeric AAV genome. The green trace was obtained with the AA5 genome sample at 5 µL with a titer of 1 x 1013 GC/mL. The intact genome (2.35 kb) as well as the small size impurities between 0.5 to 1 kb were detected. The size of short impurities in AAV5 samples was smaller than those in AAV8 samples. This might be related to the differences in production processes for making these two samples. For example, different amounts of nucleases can be used in treating the cell lysate. In addition, there can be differences in how the AAV Full capsids were collected at the end of the ultracentrifugation procedure. It is important to note that the RNA size standards migrate slower than the single stranded AAV genome of the same sizes. This is due to differences in base composition in these nucleic acid fragments as well as the differences related to ribose in RNA versus deoxyribose in single stranded DNA.
Figure 3. Analysis of AAV Samples of Different Genome Sizes.
Benefit of Benzonase Treatment: Small size impurities were present in AAV samples as shown in Figure 3. These impurities could be degraded nucleic acids from the host cell and from the plasmid vector that were not well separated from the Full AAV capsids. Benzonase is an unselective endonuclease that attacks and degrades all forms of DNA and RNA (single stranded, double stranded, linear and circular) down to oligonucleotides that are 2‒5 bases in length. An Empty AAV8 sample produced as a by-product during the production of a packaged AAV8 containing an intact genome size of 4.5 kb was obtained from Vigene. This Empty AAV8 sample at 10 µL with a titer of 1.06 x 1013 GC/mL was treated with benzonase and filtered through an Amicon Ultra-0.5 centrifugal filter unit with MW cut off of 100 kDa to remove benzonase as well as degraded nucleic acid species. The filtrate was also collected. As a control, another Empty sample at 10 µL that was not treated with benzonase was also applied to the Amicon filter. The filtrate was also collected. Since Empty AAV samples often contain a low amount of Full AAV capsids, the Empty samples were treated with Proteinase K after they were collected from the Amicon filter device. Nucleic acids. from the two Empty samples (with or without benzonase treatment) and the two filtrate samples were purified and analyzed on a PA 800 Plus System. Results in Figure 4 demonstrated that the amount of small size impurities was significantly reduced in the benzonase treated sample than in sample not treated with the enzyme. In addition, a significant amount of small size impurities was present in the filtrate from the Empty AAV sample not treated with benzonase. In contrast, much lower amount of these impurities was present in filtrate from the benzonase treated Empty AAV sample indicating that the benzonase was effective in degrading the small size impurities outside of the capsids. As expected, a small amountof intact AAV genome shown by the red arrow was detected in the two empty AAV samples regardless of benzonase treatment. However, it was not detected in the two filtrate samples.
Figure 4. Benzonase Treatment. Use of benzonase treatment led to decrease in small size impurities in an empty AAV8 sample. Sample injection: 5 kV, 3 seconds.
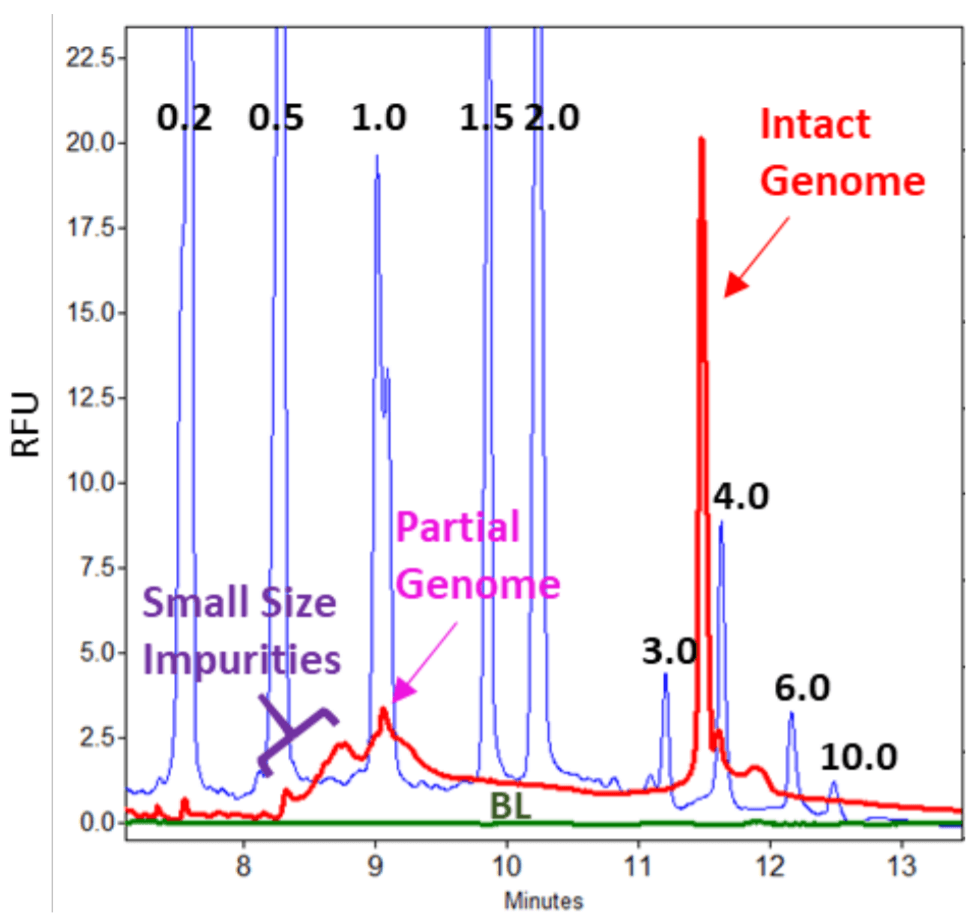
Separation of Partial AAV Genome from the Intact AAV Genome: As shown in Figure 3, the intact AAV genome and small size impurities were detected in two AAV samples with relatively smaller intact genome sizes: AAV8 (2.8 kb) and AAV5 (2.35 kb). No partial genome was detected. An AAV8 sample with a larger intact genome size (4.5 kb) was tested next. In this experiment, both Full and Empty AAV8-4.5 kb samples at 10 µL with a titer of 1.06 x 1013 GC/mL were treated with benzonase to degrade nucleic acid impurities outside of the capsids. After benzonase digestion, samples were filtered with Amicon filter, treated with protein K before nucleic acid extraction and separation on a PA 800 Plus System. Samples were injected at 5 kV for 3 seconds. As a control, another pair of Full and Empty AAV8-4.5 kb samples were processed in the same way as described above but without benzonase treatment. Results obtained with these four samples were shown in Figure 5. The peak for the intact genome was present in all 4 samples. However, the peak height was much higher in the two Full samples than in the two Empty samples. The corrected peak areas for the intact genome peak were 10944 and 9656 in Full samples treated with or without benzonase, and 571 and 573 in Empty samples treated with or without benzonase. Therefore, this analysis method has the potential to be used as a method for titer determination upon further development. In addition to the intact genome peak, the two Full samples also had a peak that is slightly larger than 1 kb. This could be the partial genome peak (1.3 kb) without the transgene. Sequencing or restriction fragment analysis will be needed to confirm this hypothesis. The small size impurity peak is minimal in both Full samples indicating that these Full samples were of high purity. As expected, the small size impurity peak in Empty sample without benzonase treatment (pink trace) was significantly higher than in the one treated with the enzyme (green trace). In Figure 2, the Full AAV 8-4.5 kb samples treated with benzonase and Proteinase K was injected at 5 kV for 10 seconds. The intact genome, partial genome and low amount of small size impurities were detected (red trace) The signal to noise ratio for the partial genome peak was 8.10. The green trace was obtained with nuclease-free water that went through the entire process of benzonase and proteinase K treatment as well as the nucleic acid purification using QIAquick PCR purification kit. Good baseline was obtained with this blank control.
Figure 5. Analysis of Full and Empty AAV8-4.5 kb Samples on a PA 800 Plus System. Samples were treated with (+B) or without (-B) benzonase, passed through Amicon filter followed by nucleic acids extraction and separation on a PA 800 Plus System. The titer for the Full and empty sample was 1.06 x 1013 GC/mL. The vertical blue bars represent the migration time positions of RNA size standards with their sizes indicated in kilobases.
Confirmation of Presence of Partial AAV Genome in Sample Not Treated with Benzonase and Filtering: The partial genome peak observed in Figure 5 was relatively small. In order to confirm this partial is a true peak, another experiment was performed with the Full AAV8-4.5 kb sample at 10 µL with a titer of 1.06 x 1013 GC/mL but without benzonase treatment and filtering. After Proteinase K treatment, the purified nucleic acids were injected on a PA 800 Plus System for 3 seconds, 10 seconds and 20 seconds, respectively at 5 kV. Results are shown in Figure 6. Both the intact genome and the partial genome peaks are present in all injections with increasing signal intensity corresponding to increasing injection time. The small size impurity peaks were also observed, which migrated between 8.5 to 9 minutes, faster than the partial genome peak which was detected around 9.2 min.
Figure 6. Analysis of Full AAV8-4.5 kb Sample with Different Electrokinetic Injection Conditions (3 to 20 Seconds). Sample was not treated with benzonase and was not filtered.
Figure 7. Standard Workflow for AAV Transgene Integrity Analysis by CE-LIF. AAV samples were treated with benzonase (B) to degrade small size impurities outside of capsids. Degraded nucleic acids were removed by filtering (F). Purified AAV capsids were treated with proteinase K (PK) to release AAV genome which was purified with QIAquick PCR purification kit (QPK) and analyzed on a PA 800 Plus System by CE-LIF using a separation gel buffer containing 1% PVP, 1x TBE, 4 M Urea and SYBR Green II.4 Blue trace: RNA size standards; Red trace: AAV genome sample extracted from packaged AAV8-4.5 kb. Green trace: blank control.
Standard Workflow for AAV Genome Integrity by CE-LIF: Based on the results shown in Figures 3 to 6, the standard workflow for AAV genome integrity analysis was established and is shown in Figure 7. Briefly, the AAV samples were first treated with benzonase. Degraded nucleic acids and lower MW impurities outside of the capsids were removed by filtering through Amicon filter with 100 kDa MW cutoff. The purified AAV capsids were treated with Proteinase K to release the viral genome. The AAV genome samples were purified using QIAquick PCR purification kit and separated on a PA 800 Plus System by CE-LIF.
Figure 8. Repeatability Test for Sample Separation. The sample prepared from AAV8-4.5 kb (10 µL with a titer of 1.06 x 1013 GC/mL) in standard workflow was injected at 5 kV for 10 seconds for 8 times.
Repeatability Test for Sample Separation: A sample prepared from AAV8-4.5 kb (10 µL with a titer of 1.06 x 1013 GC/mL) in standard workflow was injected at 5 kV for 10 seconds for 8 times. As demonstrated in Figure 8, peak patterns in 8 runs were very consistent. The CV for migration time for the partial and intact genome peaks was 0.17% and 0.30% respectively.
Figure 9. AAV Genome Size Purity Analysis Using a Simplified Workflow. The Green trace was obtained with AAV sample treated with Proteinase K. The red trace was obtained with sample applied to the Qiagen column without prior proteinase K treatment. Pink trace was obtained by digesting the filtrate from the Qiagen column with Proteinase K and then applied to another Qiagen column for purification of nucleic acids.
Quick AAV Size Purity Test Through a Simplified Workflow: Since the standard workflow takes about 3 hours to complete, a simplified workflow was tested. In this short workflow, the AAV8- 4.5 kb sample (5 µL with a titer of 1.06 x 1013 GC/mL) was not treated with benzonase or proteinase K. Instead, it was mixed thoroughly with the binding buffer from the QIAquick PCR purification kit and applied to the Qiagen column. Filtrate was collected from this sample, then, digested with proteinase K. As a control, another 5 µL sample was treated with Proteinase K before Qiagen column. Nucleic acids were purified and separated on a PA 800 Plus System by CE-LIF. Figure 9 shows that although the signal level obtained from this simplified workflow was lower than samples prepared with Proteinase K, the same peak pattern including the intact genome containing the trasngene, the partial genome and small size impurities were well conserved. It takes only about 30 minutes to test a sample from sample preparation to getting results from a PA 800 Plus System using the simplified workflow. As expected, a very low amount of intact genome was obtained from the filtrate, indicating that a very low number of capsids were not opened in the presence of high concentration of Guanidine HCl present in the Qiagen binding buffer. Similar results were also obtained with an AAV2 sample from Signagen (data not shown). This simplified workflow may be useful for quick in-process quality check during AAV manufacturing process.
Conclusions
- Capability to detect intact and partial genome helps users to get a quick assessment of the size quality of the AAV genome
- Simplified screening workflow takes 30 minutes, good forfast in-process quality check during AAV manufacturing process
- Standard comprehensive workflow provides a method for more in-depth analysis in 3 hours
- Both benzonase treatment and filtering help remove the small size impurities, simplifying data interpretation
- Methods can be applied to various AAV serotypes
- Preassembled EZ-CE cartridge saves time for users
- Urea is less toxic than denaturant used in denaturingagarose gel such as formaldehyde, sodium hydroxide and glyoxal
- CE-LIF methods generate less waste than denaturing agarose gel and southern blot
References
- Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. (2017) BioDrugs 31:317–334.
- Characterization of Genome Integrity for Oversized Recombinant AAV Vector. (2010) Molecular Therapy 18(1): 87–92.
- Viral Quantitative Capillary Electrophoresis for Counting Intact Viruses. (2011) Analytical Chemistry 83(13):5431-5.
- Method Optimization and Evaluation for RNA PurityAnalysis Using CE-LIF Technology. (2018) SCIEX Technical Notes. RUO-MKT-02-8017-B
Acknowledgements
We thank Andras Guttman, Marcia Santos, Mukesh Malik, and Elliott Jones for helpful discussions.
Request a full version by clicking the link below:
https://info.sciex.com/genome-integrity-analysis-aav
 Click to enlarge
Click to enlarge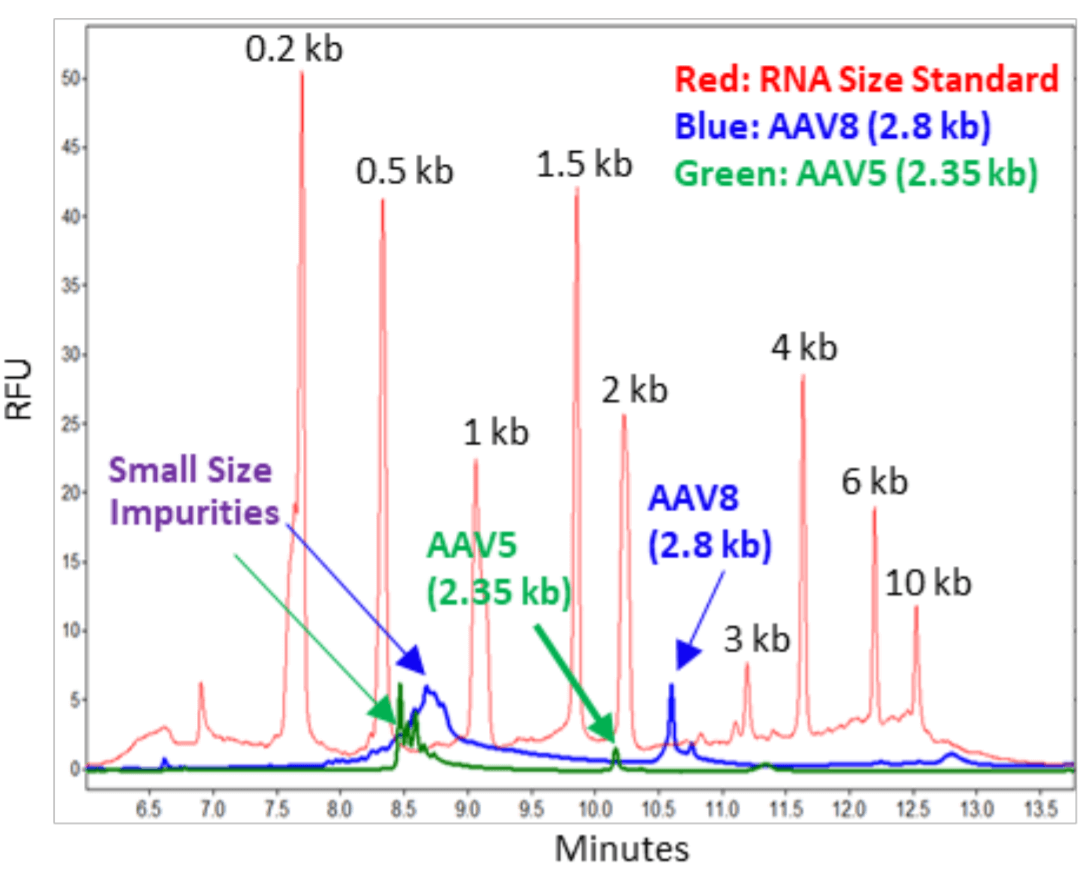 Click to enlarge
Click to enlarge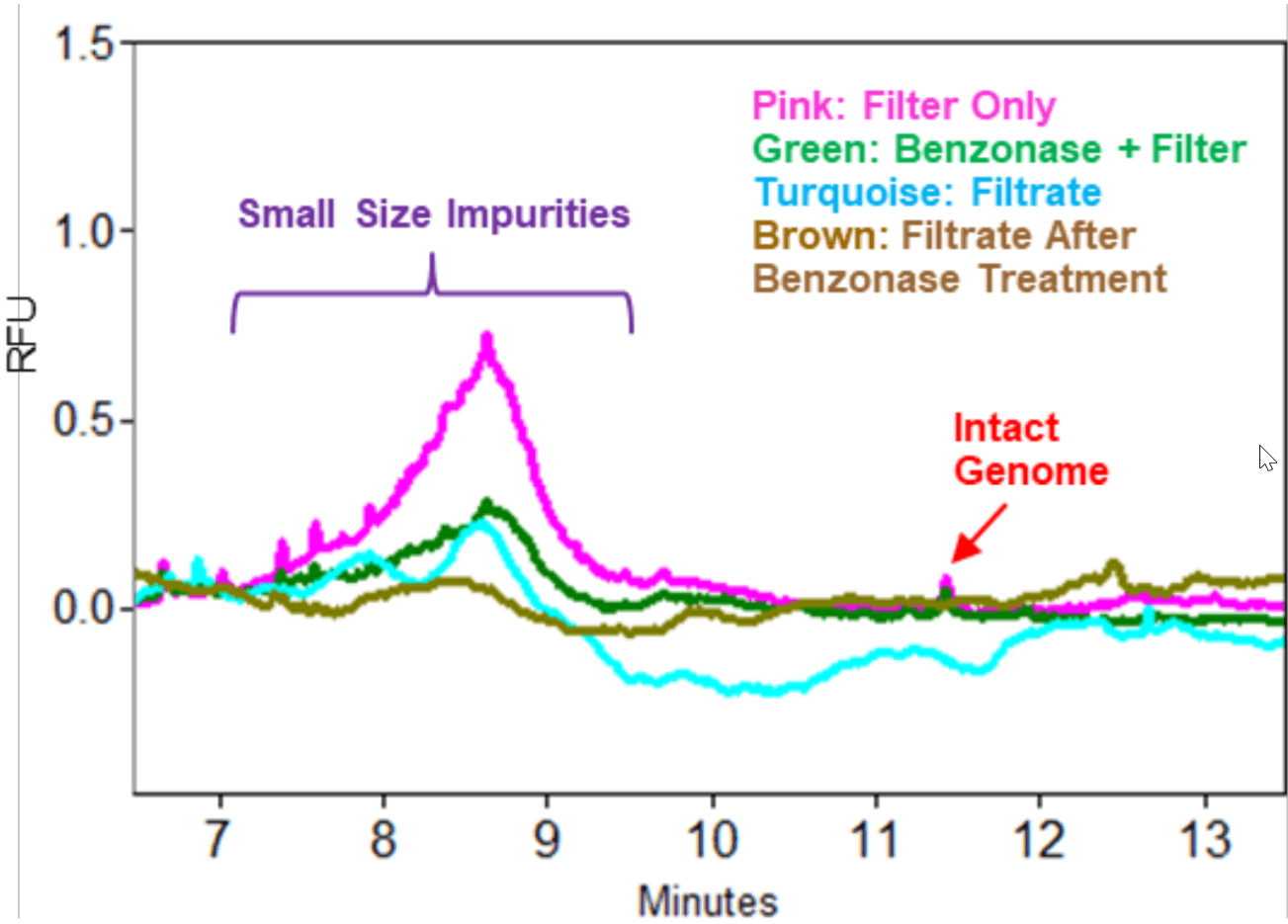 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge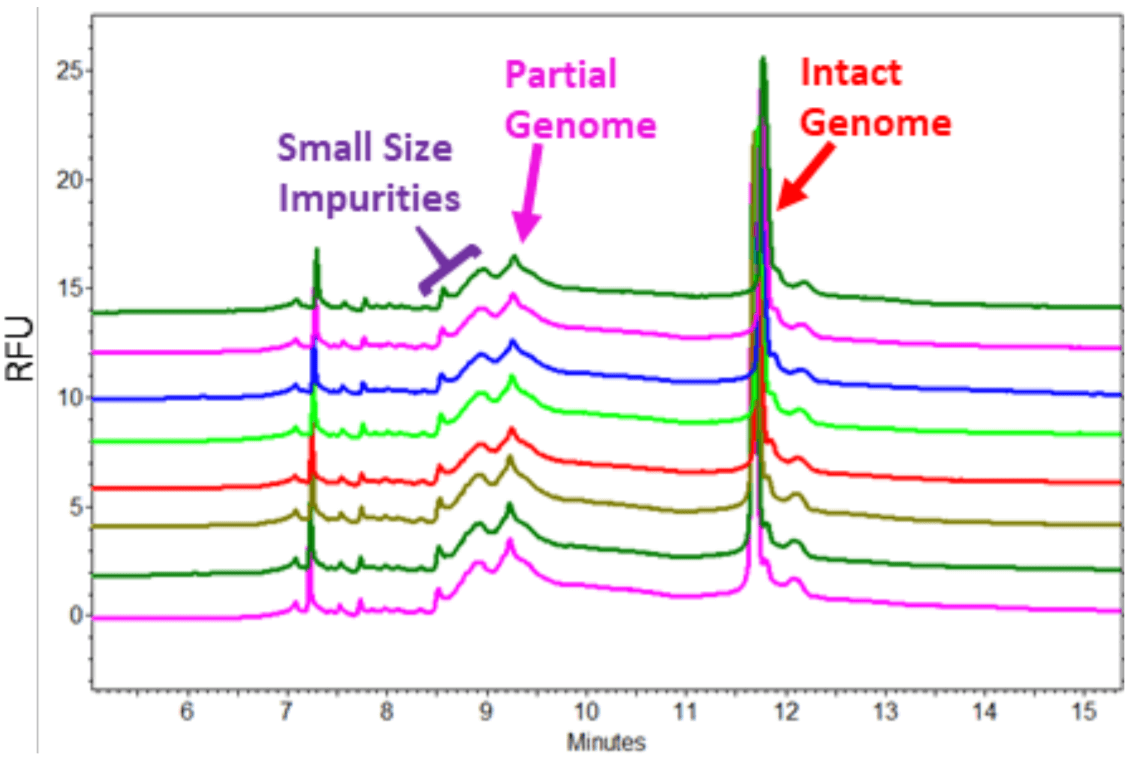 Click to enlarge
Click to enlarge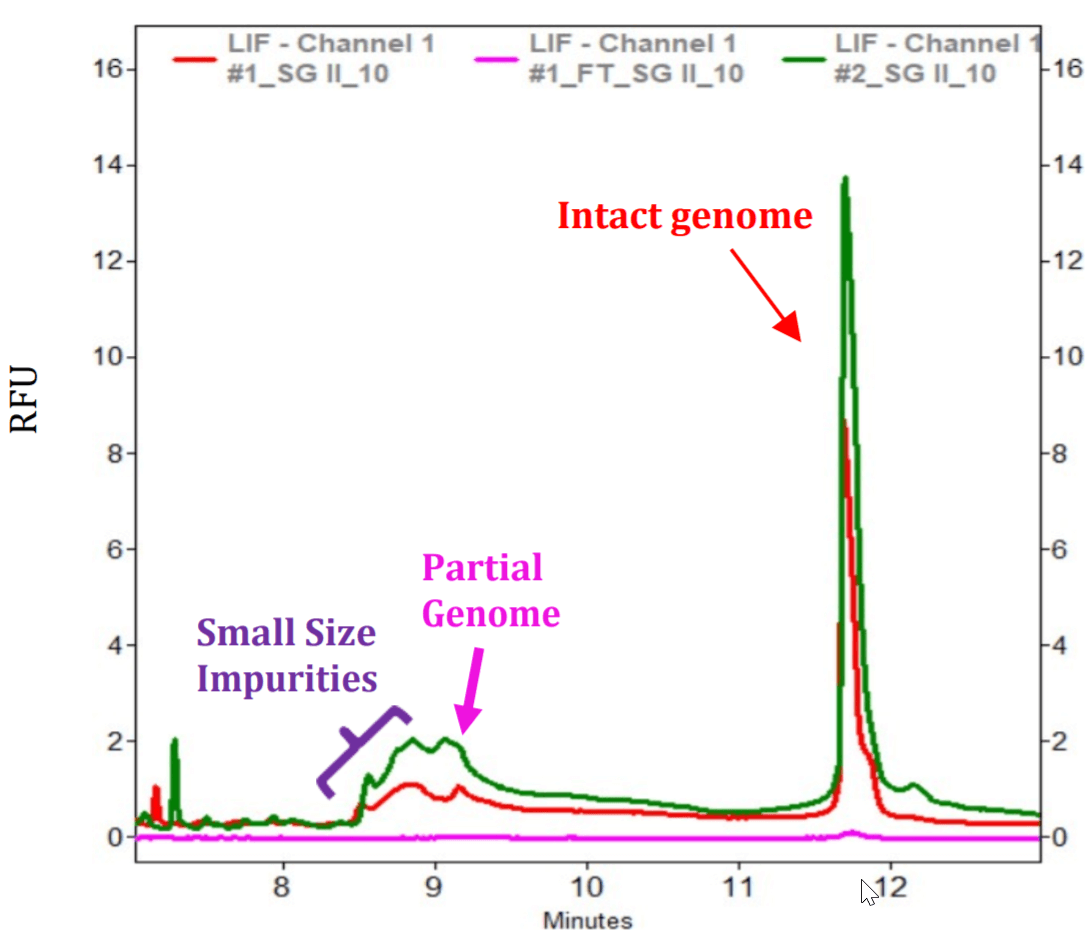 Click to enlarge
Click to enlarge