Sequencing Analysis as A Tool for Fast and Accurate Microbial Contaminants Identification
• High Specificity • Rapid Results • Superior Over Conventional Microbial Methods
J. Luo1, K. Uchiumi1 , M. Souquet2 and H.Yowanto1
1 SCIEX, Brea, California, USA
2 SCIEX Darmstadt, Germany
Introduction and Overview
During the development and manufacturing processes of biologics, it is critical to monitor the sterility of pharmaceutical ingredients, water for pharmaceutical use, the manufacturing environment, intermediates and finished products. In addition, Master Cell Bank, each seed lot and cells used in each production run should be tested for adventitious agents including mycoplasma, bacteria, fungi, viruses, and virus-like particles. When microbial growth is observed, an investigation needs to be conducted and FDA guideline requires sterility test isolates be identified to the species level. It has been shown that DNA sequencing is superior in identifying microbes to the species level, when compared against conventional microbiology staining and culture methods. In this poster, we describe a process for sequencing ribosomal RNA genes of microorganisms that offers fast turnaround time (8 hrs) and high microbial identification accuracy. Upon isolation of nucleic acid from each microbial sample, the gene target is amplified using a polymerase chain reaction (PCR), followed by sequencing reaction using the Dye Terminator Cycle Sequencing (DTCS) on the GenomeLab GeXP Genetic Analysis System. Results are compared against the NCBI microbial library to identify the contaminants.
Methods
Genomic DNA Isolation: One loopful of bacteria sample from a single colony was re-suspended in 100 µl of sterile water in a microcentrifuge tube. The tube was placed in a boiling water bath for 10 minutes. After a minute spin at 10,000 g, 5 ul of the supernatant containing microbial DNA was used as the PCR template. For yeast sample, Genomic DNA was extracted using Beckman Coulter Agencourt Genfind V2 kit with a lysis buffer that contained lysozyme and Lyticase from Sigma.
Target Gene Amplification: For Clostridium species and yeast contaminants, each PCR reaction (50 µl total volume) contained 5 µl of sample genomic DNA, 1.5 mM MgCl2 , 200 nM dNTPs, 200 nM Primers and 0.04 units of AmpliTaq Gold. Cycling conditions were: 95° C for 5 min followed by 30 cycles of 95° C for 30 sec, 50° C for 30 sec and 72° C for 1 min; followed by a final extension at 72° C for 10 min and 4° C forever. For non-pathogenic bacteria contaminant, the Takara RR180A kit was used following the manufacturer’s instructions. Thermal cycling conditions were: 94° C, 1 min followed by 30 cycles of 94° C for 30 sec, 55° C for 30 sec and 72° C for 1 min; followed by a final extension at 72° C for 3 min and 4° C forever.
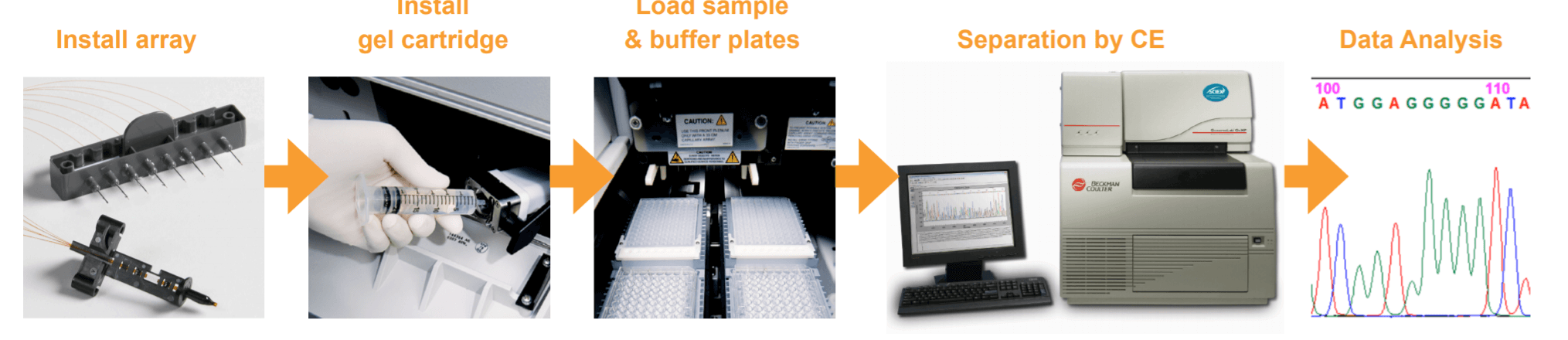
Figure 1. Basic operator steps for using the GenomeLab GeXP Genetic Analysis System.
Post PCR Clean-up: Agencourt AMPure XP from Beckman Coulter was used for post PCR clean-up. The process of Agencourt AMPure XP (Figure 2) takes less than 15 minutes to complete.
Sequencing and Data Analysis: Sequencing reactions were set up per instructions for the DTCS Quick Start Kit from SCIEX. Separation and data analysis are fully automated on the GenomeLab GeXP Genetic Analysis System. Sequences were submitted to NCBI Database for BLAST search to identify the contaminant.
Figure 2. Post PCR clean-up process using Agencourt AMPure XP.
Figure 3. Sanger Dye Terminator Cycle Sequencing.
Figure 4. Work Flow for Microbial Identification using GenomeLab GeXP Genetic Analysis System.
Results
Identification of Clostridium septicum in a sample. The 16s rDNA PCR product was sequenced using DTCS Quick Start Kit. Sequencing fragments were purified by ethanol precipitation and separated on a GeXP instrument. Figure 5 shows the sequencing results. After automated quality trimming, a 497bp sequence was submitted for BLAST search against the 16s rRNA database at the NCBI website. Alignment results (Figure 6) indicated that the query sequence was 100% identical to 16s rDNA from Clostridium septicum.
Figure 5. Sequencing results showing raw and analyzed data as well as quality values and DNA sequence.
Figure 6. BLAST search results obtained with a 497 bp 16s rDNA sequence in NCBI website.
Identification of E. coli in a sample. A non-pathogenic bacteria sample was obtained from Takara, Japan. Genomic DNA was extracted by a quick cell lysis in a boiling water bath. Full length 16s rDNA was amplified and sequenced using DTCS Quick Start Kit and GeXP System. Consensus sequence was created in Figure 6. BLAST search results obtained with a 497 bp 16s rDNA sequence in NCBI website. p4 Gene Studio (Figure 7) and submitted to NCBI for BLAST search. Search results (Figure 8) showed the query sequence had high homology to pathogenic bacteria and non-pathogenic E. coli. Since the sample was known to be non-pathogenic, the sample was identified as an E. coli species.
Figure 7. Contig Assembly in Gene Studio to create consensus sequence from forward and reverse sequencing results.
Figure 8. BLAST search results with a 1443bp 16s rDNA sequence from Sample#1. Sequences with the highest homology to the query sequence are 16s rDNA from pathogenic bacteria (Escherichia Fergusonii; Escherichia Albertii; Escherichia coli O157 and Shigella species) and non-pathogenic bacteria (Escherichia coli NBRC 102203, Escherichia coli strain K12 and Escherichia coli Strain U5/41). Since the sample is non-pathogenic, the sample was identified as an E. coli species.
Identification of a Wild Yeast 2 contaminant in a beverage sample. The microbial contaminant was initially typed as Candida krusei (Wild Yeast 1) by culture method by a commercial vendor. The 18s rDNA was sequenced using DTCS Quick Start Kit and GeXP System. After quality based trimming, the 18s rDNA sequence (Figure 9) was submitted to NCBI for BLAST search. The contaminant was identified as Candida tropicalis (Wild Yeast 2).
Figure 9. The 18s rDNA sequence of the beverage contaminant and its alignment results in NCBI.
 Click to enlarge
Click to enlarge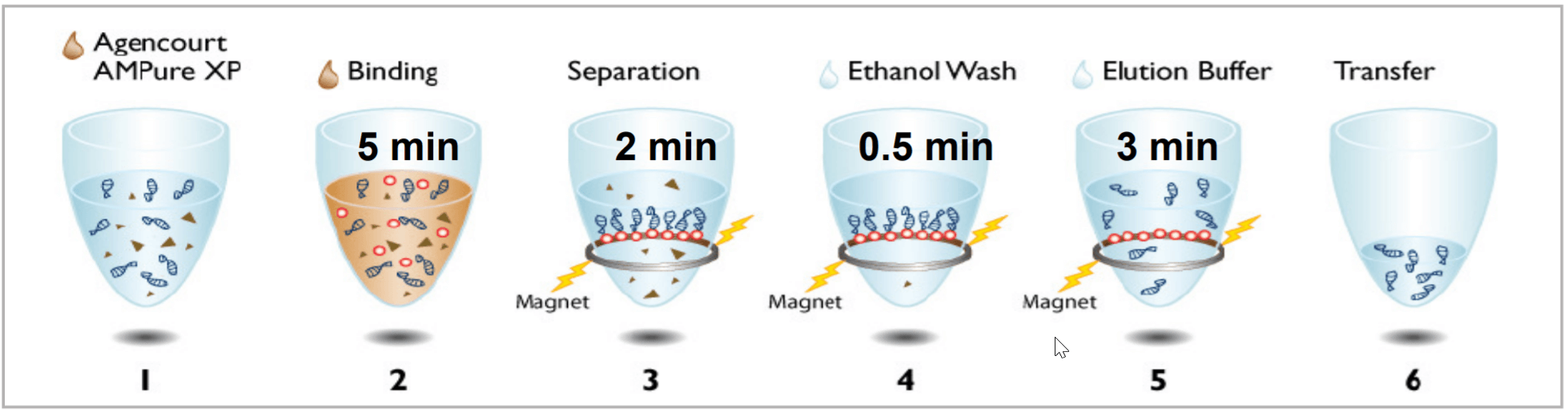 Click to enlarge
Click to enlarge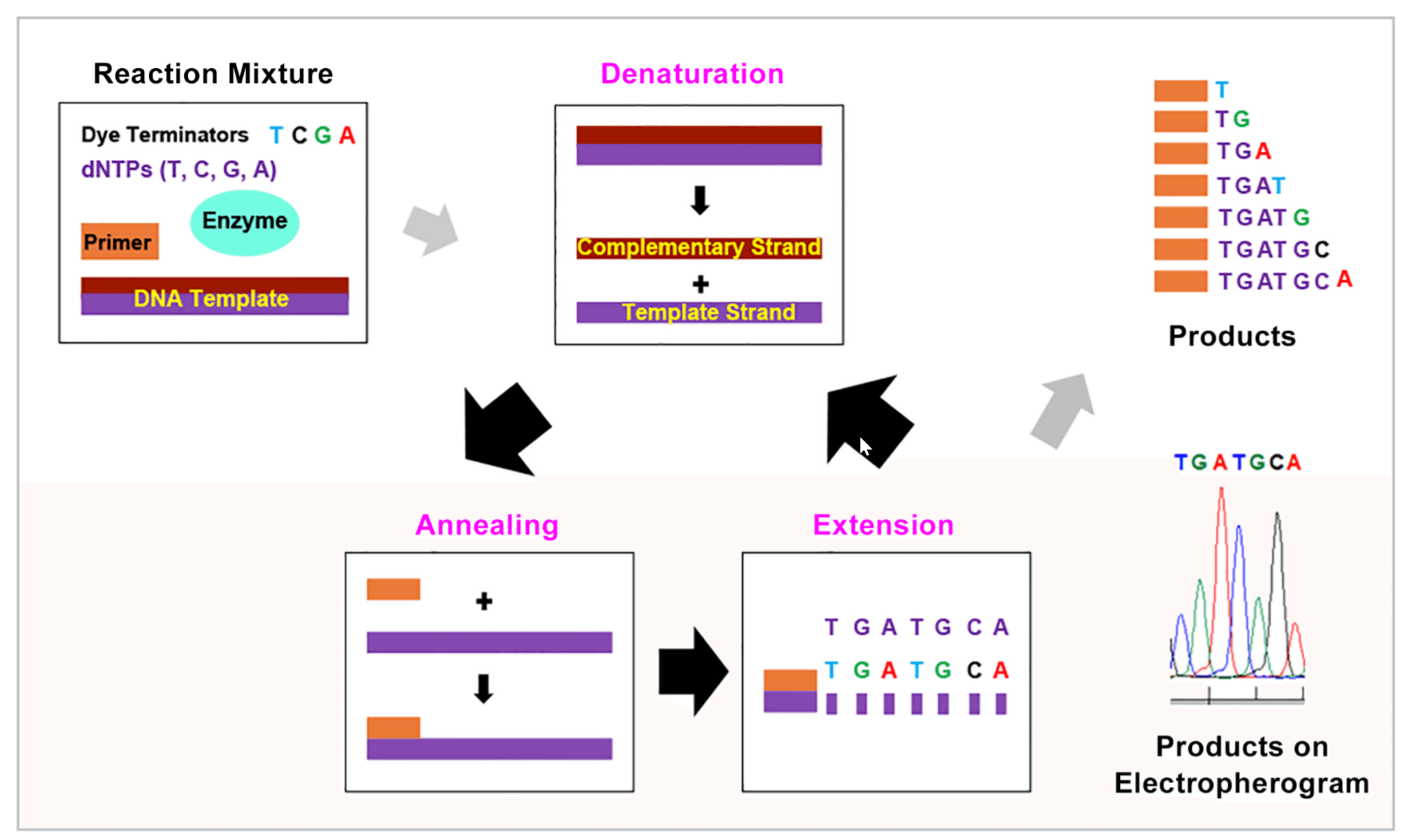 Click to enlarge
Click to enlarge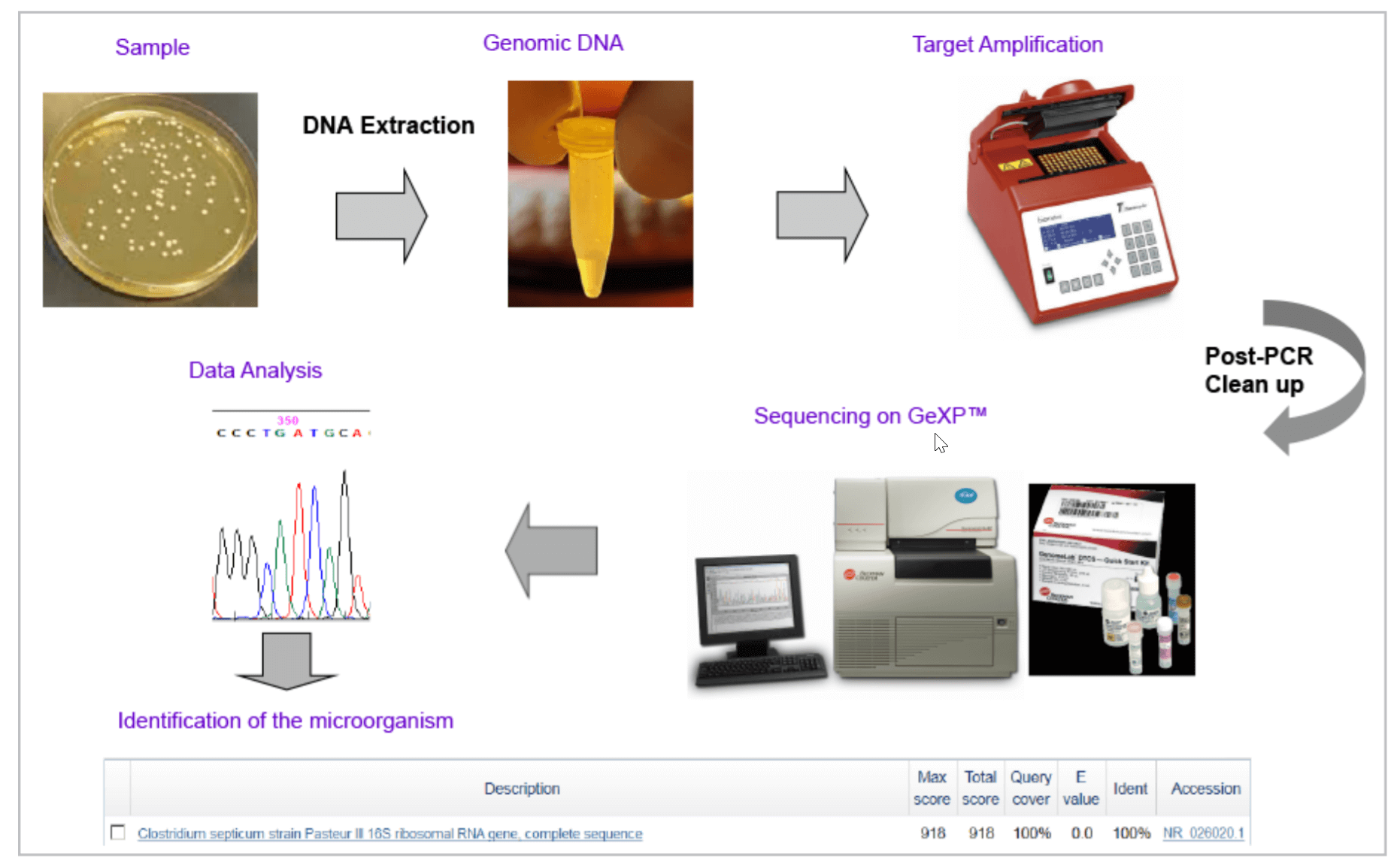 Click to enlarge
Click to enlarge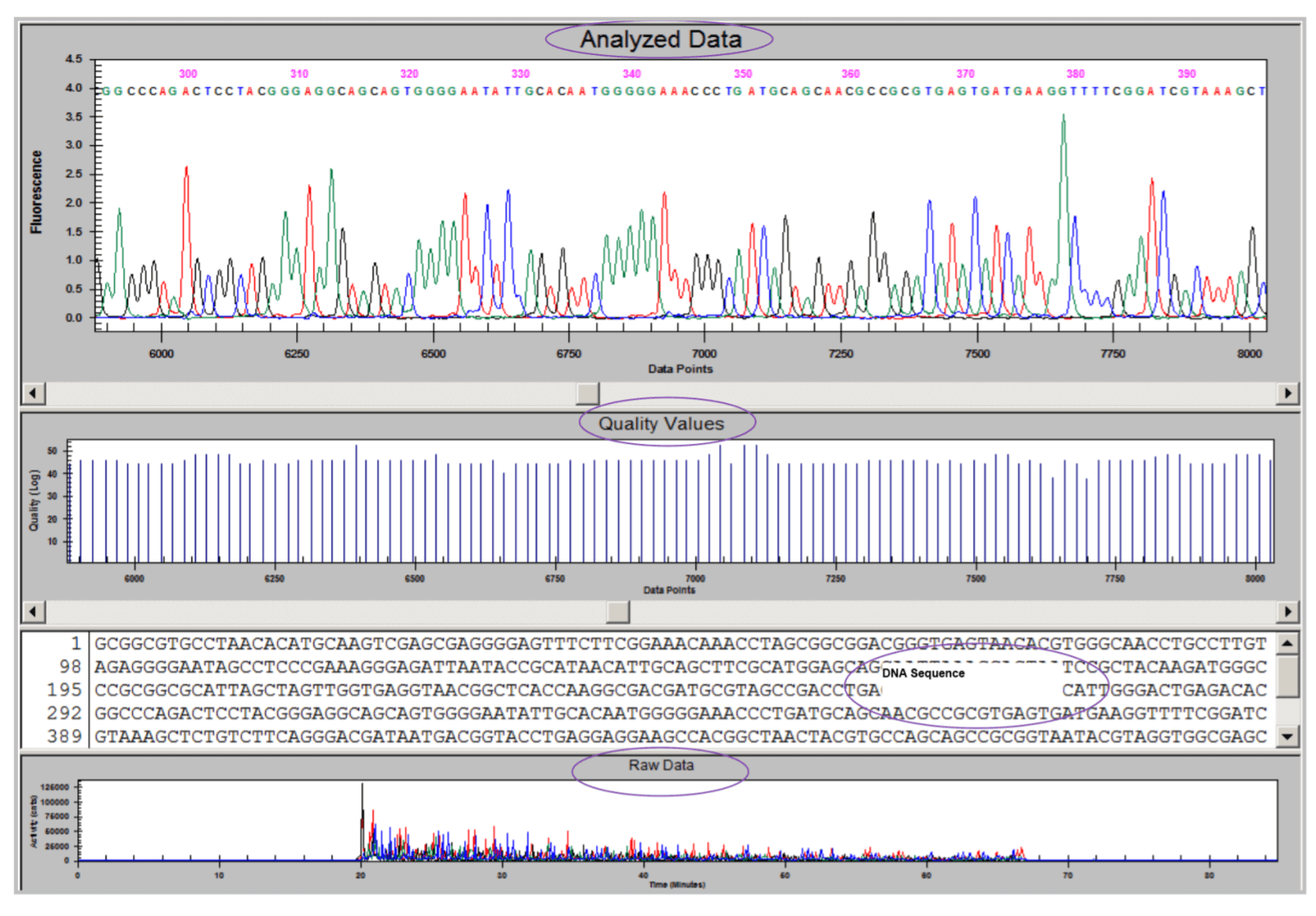 Click to enlarge
Click to enlarge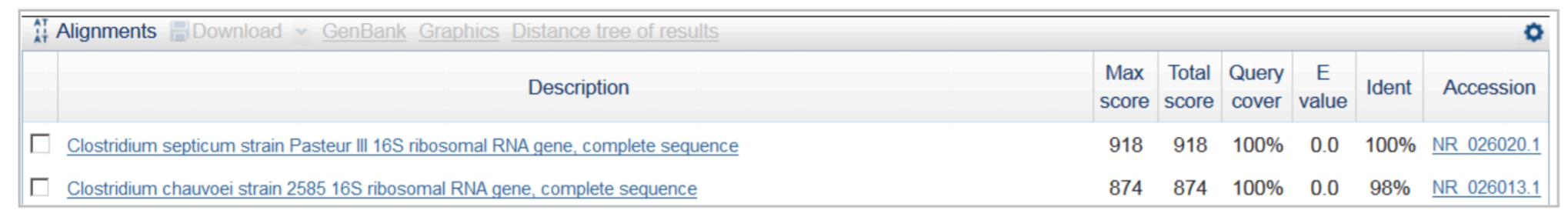 Click to enlarge
Click to enlarge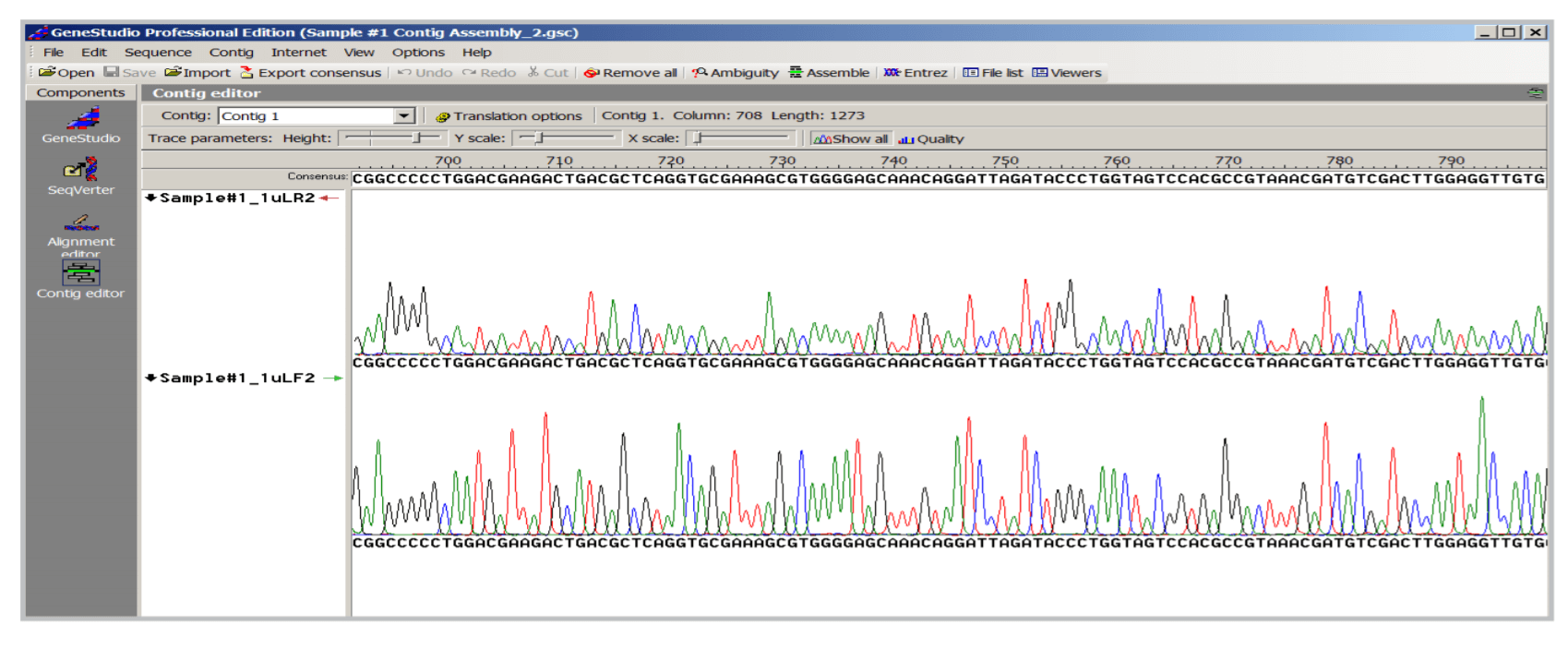 Click to enlarge
Click to enlarge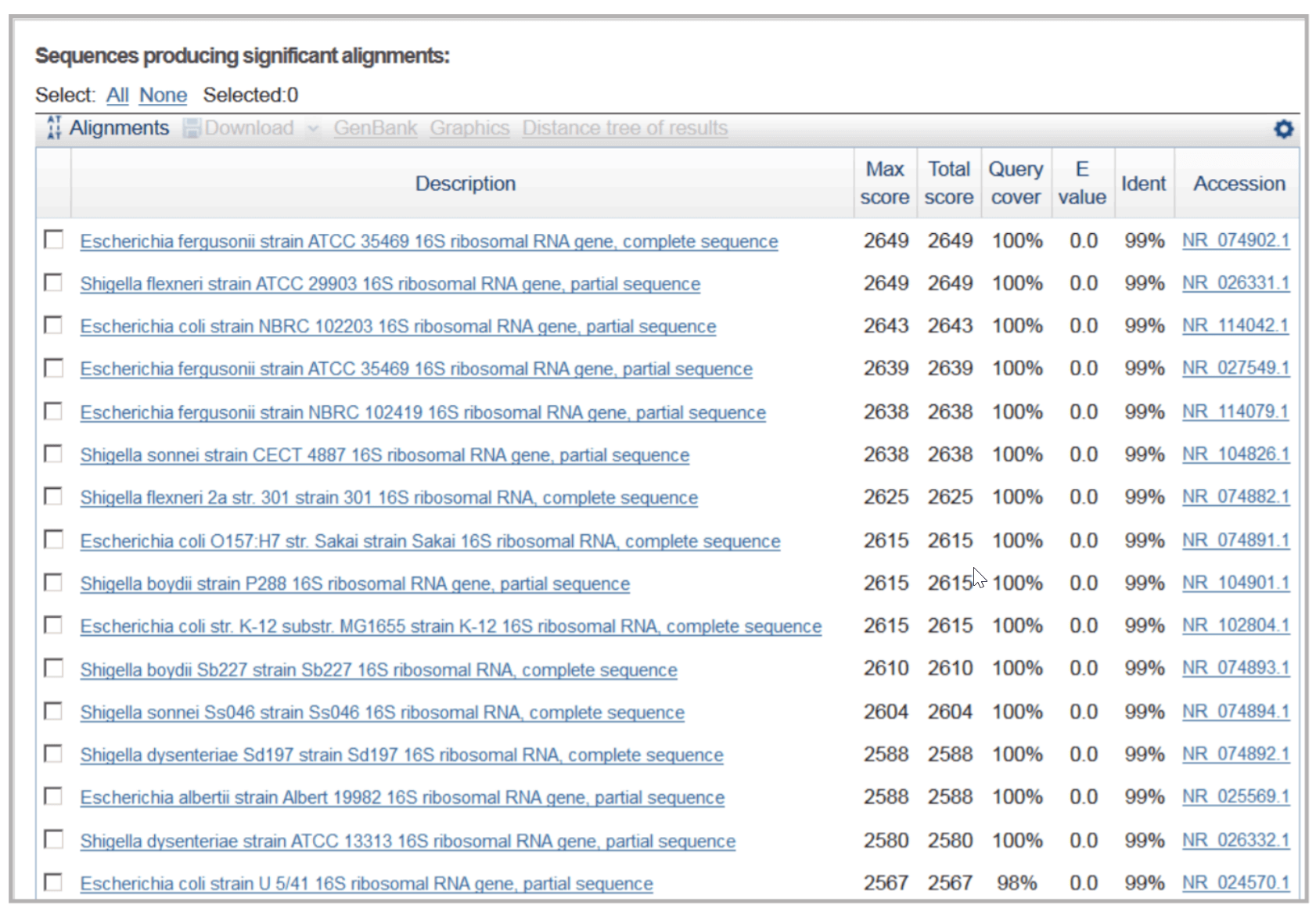 Click to enlarge
Click to enlarge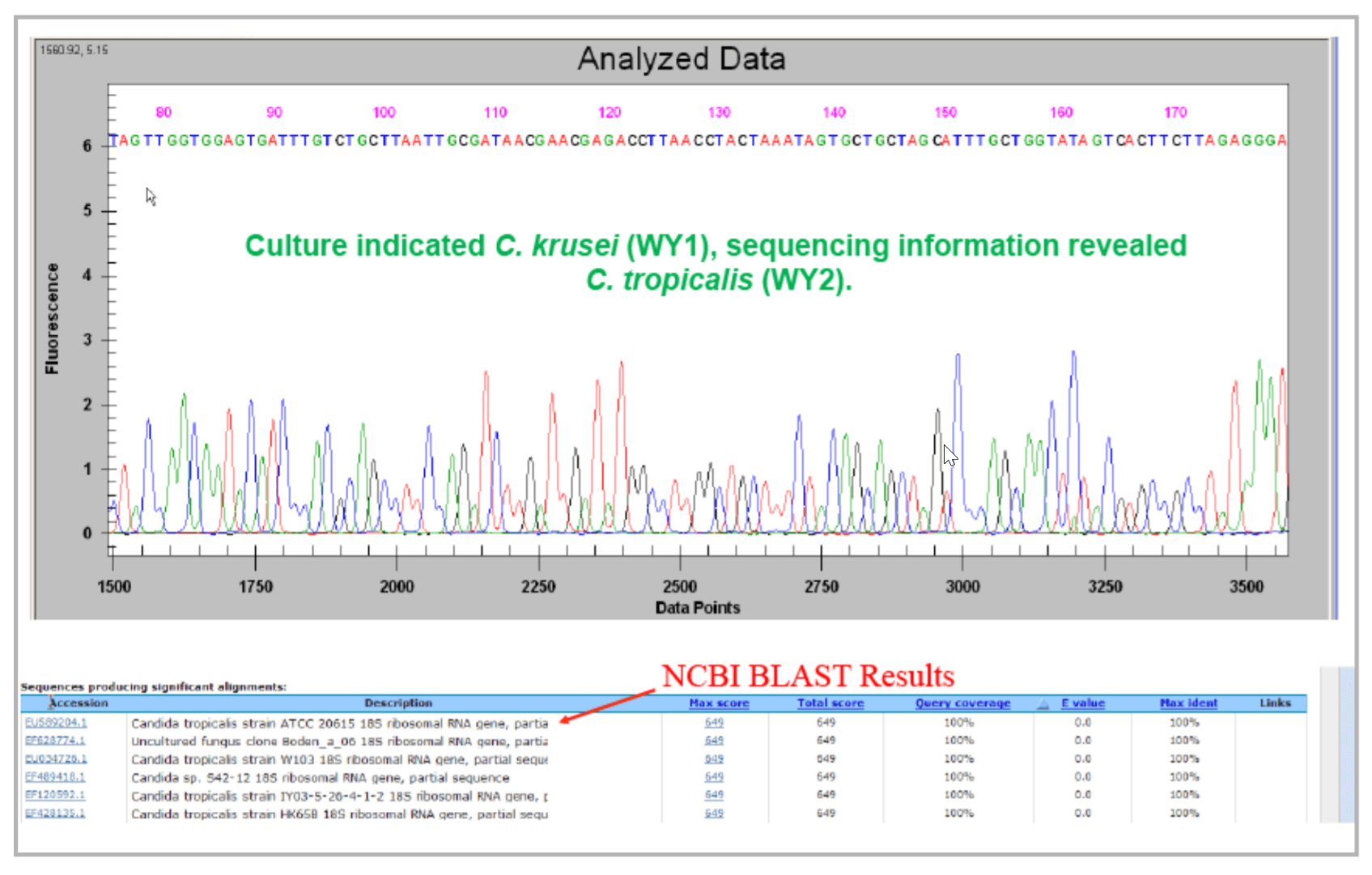 Click to enlarge
Click to enlarge