Quantitation and identification of the pesticide malathion in fruit samples using MRM3 workflow
Using the QTRAP® 5500 System
André Schreiber, Tania Sasaki, Tanya Gamble
SCIEX, USA
Abstract
MRM and MRM3 quantitation organophosphorus pesticide Malathion in fruit samples using the QTRAP® 5500 System. Higher selectivity of MRM3 eliminates matrix interference for much better sensitivity, accuracy, and reproducibility for the qualifier signal in fruit matrix. MRM3 gave comparable data versus MRM for the quantifier signal.

Introduction
LC-MS/MS instruments operating in Multiple Reaction Monitoring (MRM) are widely used for targeted quantitation on triple quadrupole and hybrid triple quadrupole linear ion trap systems (QTRAP® Systems) because of their well-known selectivity and sensitivity. Although this double mass filtering greatly reduces noise there is always a chance that elevated background levels or matrix signals interfere with the targeted analyte.
One possibility of improving quantitative results is to use a more selective detection mode, such as MRM3 workflow.1 In comparison to MRM mode, MRM3 often provides higher selectivity due to one additional fragmentation step, effectively reducing interferences and improving quantitation.
Improved selectivity for quantitation
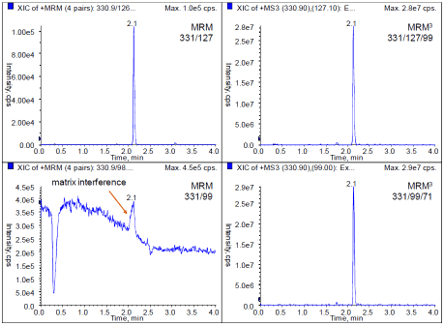
Here, a homogenized apple was spiked with the organophosphorus pesticide Malathion (10 ppb), then extracted using a QuEChERS procedure, diluted 50 times to minimize matrix effects, and analyzed by LC-MS/MS. The resulting chromatograms using two MRM transitions and two MRM3 experiments are shown in Figure 1. The MRM transition 331/127 showed expected selectivity, while the second transition 331/99 had an elevated background level and also matrix interference. In contrast both MRM3 experiments showed superior selectivity for reliable quantitation. Table 1 shows signal-to-noise ratios (S/N) of the chromatograms described above.
Figure 1: Comparison of MRM vs MRM3 for detecting malathion (10ppb) in apple extract. (Left) While the MRM transition 331/127 showed expected selectivity, the second transition 331/99 had an elevated background level and also matrix interference. This results in incorrect ion ratios and prevents confident identification. (Right) In contrast, both MRM3 experiments showed minimal interferences and higher selectivity.
Improved confirmation
The quantifier MRM and the quantifier MRM3 had very similar sensitivity. Both experiments allowed quantifying Malathion at sub ppb levels. However, pesticide testing requires identification. Thus, a second MRM or MRM3 signal has to be recorded to allow ratio calculation (qualifier/quantifier). In this example, the second MRM showed a dramatic loss in S/N because of the elevated background. In this case the quantitation and identification of Malathion in fruit matrix using MRM3 was much more sensitive than in MRM mode.
Methods
Chromatography: Separation was performed using a Shimadzu UFLCXR system with a Phenomenex Synergi Fusion-RP (2.5 μm) column and a fast gradient of water and methanol with ammonium formate buffer.
Mass spectrometry: A looped MS experiment was performed using the SCIEX QTRAP 5500 System with Turbo V™ Source and ESI probe. The first experiment consisted of two MRM transitions of 100 msec dwell time each. The second experiment was two MRM3 scans using a scan speed of 20000 Da/s with 20 msec fill time and 25 msec excitation time. Total cycle time of only 0.33 sec which allowed ~15 data points across the UHPLC peaks that had a base-to-base peak width of only 5 sec. The detected masses in MRM and MRM3 mode with compound dependent parameters are shown in Figure 2.
Data processing: Data was processed with MultiQuant™ Software.
Figure 2. Developing the MRM and MRM3 workflow methods. (Top) Full scan MS/MS of Malathion shows two fragments at m/z 127 and 99 for use in the MRM experiment. (Middle) MS/MS/MS of 331/127 shows a fragment at m/z 99 for use in the first MRM3 experiment. (Bottom) MS/MS/MS of 331/99 shows a fragment at m/z 71 for use in the second MRM3 experiment. |
Table 1. Improvements in detection Using MRM3 Workflow. Signal-to-noise ratios (S/N) of MRM and MRM3 of 10 ppb Malathion in a 50-times diluted QuEChERS extract of apple.
Improved reproducibility
Several fruit samples were fortified with 10 ppb Malathion and analyzed in replicates. While MRM detection suffered from matrix interference, MRM3 resulted in more accurate and reproducible data. The %CV values in apricot, apple, pear, and orange were <5% with accuracies ranging from 90% to 110%. In addition, the MRM3 ratio calculation clearly identified the presence of Malathion in the analyzed fruits.
Conclusions
LC-MS/MS methods were developed to quantify and identify the organophosphorus pesticide Malathion in fruit samples using the QTRAP® 5500 System. The traditional MRM and MRM3 modes were compared regarding selectivity, sensitivity, accuracy and reproducibility. The results show that the higher selectivity of MRM3 eliminates background and matrix interference, resulting in better data quality. MRM3 gave comparable data versus MRM for the quantifier signal but much better sensitivity, accuracy, and reproducibility for the qualifier signal in fruit matrix.
References
- MRM3 quantitation for highest selectivity in complex matrices. SCIEX technical note RUO-MKT-02-2739-A.
 Click to enlarge
Click to enlarge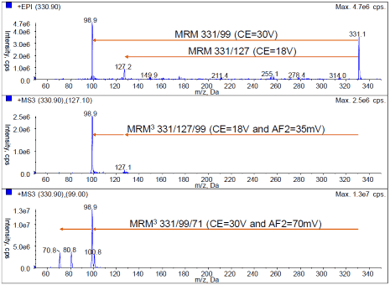 Click to enlarge
Click to enlarge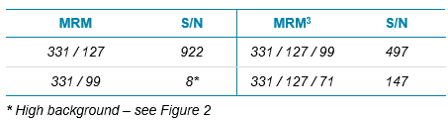 Click to enlarge
Click to enlarge