The inherent complexity of proteomes, with their variability and dynamic range, have dictated the need to maximize discovery and quantification capabilities without compromising one over the other.
In life science research, “shotgun” or bottom-up proteomics (tandem LC-MS/MS) is a key method used to comprehensively analyze proteins and their changes - including post-translational modifications (PTMs) - to give a global interpretation of biological systems. In general, there are two main approaches to generate tandem mass spectra, both which are supported by SCIEX solutions including: data dependent acquisition (DDA), also known as information dependent acquisition (IDA), and SWATH data independent acquisition (DIA).

Immerse yourself into the depths of the proteome
Proteomics research
Confidence from discovery to translational research and beyond
Overview
In pursuit of nanoscopic discoveries for big world impact
Data/information dependent acquisition (DDA/IDA)
The speed and sensitivity of the ZenoTOF 7600 system, which uses using the Zeno trap at scan speeds in excess of >133 Hz, delivers the highest-quality MS/MS data during data-dependent acquisition (DDA), even on low-abundance proteins.
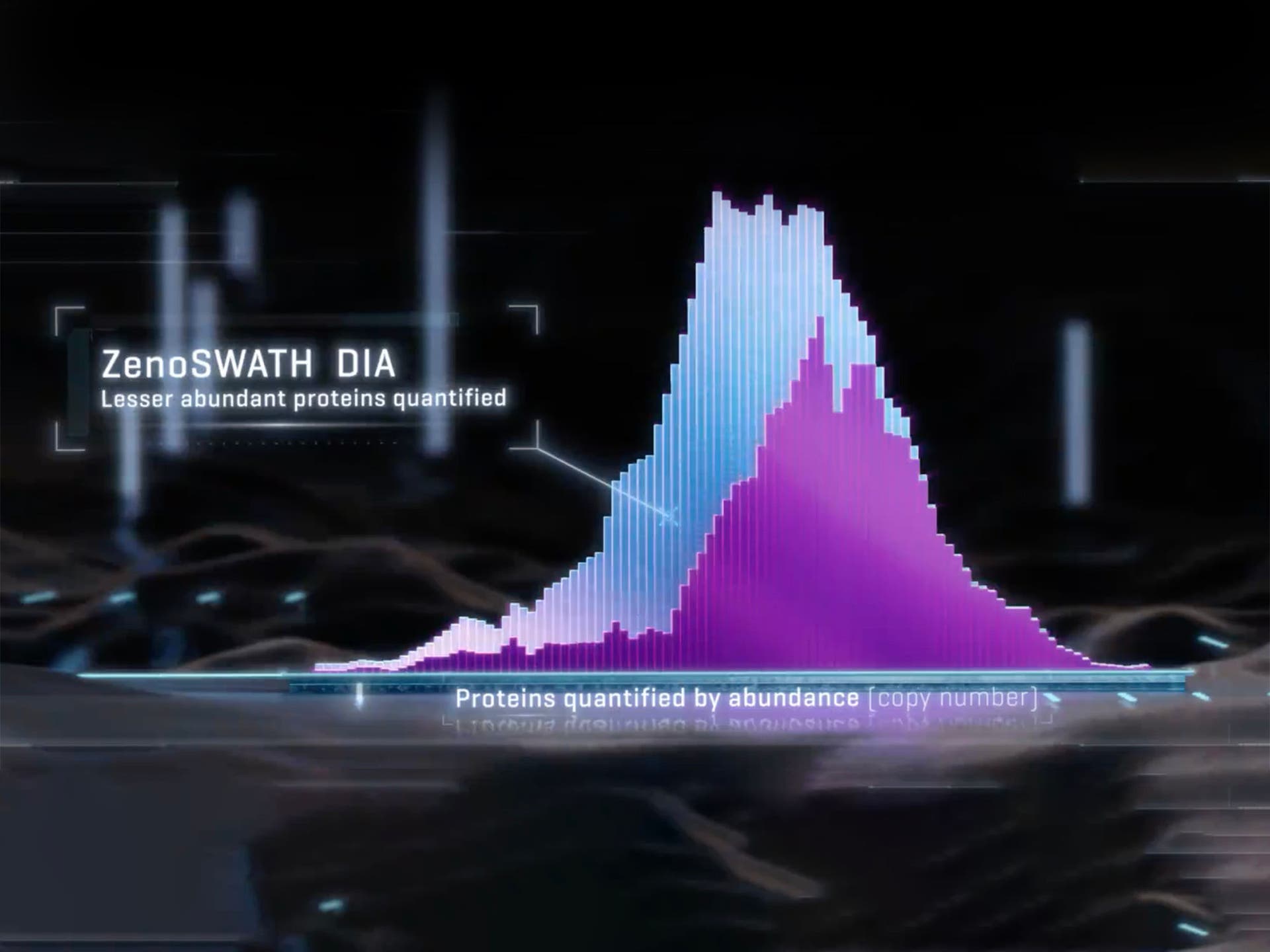
SWATH data independent acquisition (DIA)
SWATH DIA delivers precise, extensive, label-free quantification that drives biomarker discovery and identification of differentially expressed proteins. The ability to generate MS/MS data on all observable species at speed enables this approach to simultaneously identify and quantify thousands of proteins, even during short run times and with small sample loads.
Discovery proteomics
Confident identification and quantification
Move research from discovery to translational, and beyond, with more confident and significant biomarker signatures to uncover insights in their biology and mechanism of action. Using the latest technologies from SCIEX, a whole new level of proteomic depth can be discovered by uncovering unique fragmentation patterns and low abundant species.
Next-generation proteomics
Protein profiling and characterization
From post-translational modifications (PTMs) to full proteome profiling and characterization, the dynamic nature of proteins make them very challenging to analyze. Researchers require advanced technology to study protein dynamics reliably and reproducibly to determine profile protein structure changes, including PTMs profiling. SCIEX solutions allow researchers to reliably and reproducibly profile protein structure changes using the latest innovations in fragmentation technology to yield rich and informative MS/MS. Benefit from 100% sequence coverage in a single injection with and identify low-level PTMs faster than ever before.
Targeted proteomics
Monitor and quantify proteins of interest with absolute precision
Verification and validation of protein biomarkers require precise, targeted quantification across large numbers of samples to enable translation into clinical research. SCIEX solutions for targeted proteomics are driven by renown MS/MS sensitivity and precision further enhanced by Zeno trap and Zeno SWATH DIA technology.
Discover our range of proteomics solutions
Extraordinary discoveries demand extraordinary proof. The SCIEX ZenoTOF 8600 system combines proven technology from our most sensitive triple quad with that of our most versatile Zeno trap-enabled QTOF. Delivering up to 10x improvements in sensitivity* means achieving lower limits of quantitation and enabling enhanced performance across a multitude of accurate mass workflows.
* Compared to the ZenoTOF 7600+ system
Continuing the (r)evolution and taking biology beyond the ID numbers. Built off the ZenoTOF 7600 system and engineered with added specificity thanks to the scanning quadrupole dimension, the ZenoTOF 7600+ system delivers enhanced speed, depth, and certainty in quantitative measurements. Enabled with both ZenoSWATH DIA and ZT Scan DIA, the ZenoTOF 7600+ system is Ideal for high-throughput workflows and those with low sample volumes.
The instrument that began the Zeno revolution.nThe ZenoTOF 7600 is a high-resolution mass spectrometry solution driven by the power of the Zeno trap coupled with tunable electron-activated dissociation (EAD) fragmentation technology. Your workhorse for everything from untargeted to comprehensive quantitative analyses to in-depth structural characterization.
Setting a new standard for instrument resilience and robustness, the SCIEX 7500+ system gives you the confidence to face evolving analytical challenges. Take on tight timelines with exceptional sensitivity and with the power to control your instrument downtime.
Streamline your targeted proteomics workflows with the usability, efficiency and integrity of SCIEX OS software.
Simple, plug-and-play nanoflow capability with cutting edge features maintains the perfect balance of robustness and sensitivity.
Cloud-based software to extract and visualize mass spectrometry-generated proteomics data.
Proteomics resources
-
Technical note
Going library-free for protein identification using Zeno SWATH DIA and in silico-generated spectral libraries
This Zeno SWATH DIA workflow, using an in silico-generated library processing with DIA-NN software, was superior to the traditional shotgun proteomics approach using Zeno DDA for the identification of large numbers of proteins from complex samples, and agreed with two commonly used experimentally generated libraries.
-
Technical note
Zeno MS/MS with microflow chromatography powers the Zeno SWATH DIA workflow to quantify more proteins
As quantitative proteomics evolves, larger biological cohorts are being studied, often with precious samples. This creates the need to acquire data faster and with smaller amounts. Zeno SWATH DIA, with Zeno MS/MS and microflow LC, has the potential to significantly enhance core quantitative proteomics workflows.
-
Technical note
High-throughput quantitative proteomics using Zeno SWATH DIA and the Evosep One system
Proteins and peptides are important in translational research to understand biological function. This method achieves 5-6x more MS/MS sensitivity, resulting in this DIA approach surpassing DDA for protein identifications and quantification in complex matrices.
-
Technical note
Protein quantification at subnanogram loads using Zeno SWATH DIA and nanoflow chromatography
DIA has emerged as a comprehensive workflow for label-free quantitative proteomics, allowing for the acquisition of MS/MS spectra on all detectable peptides. Zeno SWATH DIA in the ZenoTOF 7600 system leverages Zeno trap activation for 5-6x increases in peptide MS/MS sensitivity in SWATH DIA workflows.
-
Technical note
Large scale protein identification using microflow chromatography on the ZenoTOF 7600 system
Generation of spectral libraries is often thought of as a time consuming procedure, but in reality, they can be generated quickly. Using microflow chromatography with fast MS/MS acquisition on a QTOF, then searching the data in the cloud can enable library generation in <48 hrs.
-
Technical note
Nanoflow Zeno SWATH DIA for high-sensitivity protein identification and quantification
Nanoflow chromatography is used in workflows to obtain the highest sensitivity. High-quality chromatographic separations are important, as good peak shape and peak resolution can reduce ion suppression and allow the MS system to sample as many unique peptides as possible.
-
Technical note
Large-scale, targeted, peptide quantification of 804 peptides with high reproducibility, using Zeno MS/MS
A highly multiplex targeted peptide quantification assay has been developed to explore the quantitative capability of Zeno MS/MS. Using a mixture of 804 heavy labeled synthetic peptides dosed into plasma, the increase in peptide sensitivity due to Zeno MS/MS was found to be 5.6 fold.
-
Technical note
PTM site localization and isomer differentiation of phosphorylated peptides
Specific location of phosphorylation sites on proteins is important in fully understanding their role in cellular processes. Here, the use and benefits of tunable electron activated dissociation (EAD) for phosphopeptide analysis and confident site-localization was evaluated on the ZenoTOF 7600 system.
-
Technical note
Tunable electron activated dissociation (EAD) MS/MS to preserve particularly labile PTMs
Determining the identity and the precise location of a post-translational modification (PTM) on a protein is important to fully characterize function. Here, the utility of electron activated dissociation on the SCIEX ZenoTOF 7600 system was investigated for the characterization of labile PTMs.
-
Technical note
Reproducible targeted peptide profiling using highly multiplexed MRM assays
Using the scheduled MRM algorithm to build highly multiplexed peptide MRM methods enables a higher number of MRMs to be monitored concurrently while maintaining optimal dwell and cycle times. The 7500 system for large scale targeted peptide quantification was evaluated using microflow chromatography.
-
Technical note
Improved LC-MRM quantification sensitivity for cyclic peptides from the natriuretic peptide family
Cyclic peptides have been identified as important therapeutic modalities. This is related to their stability in blood and their potential for oral dosing. LC-MS method development to quantify trace levels of cyclic peptides in biological matrices has remained challenging.
-
Technical note
Enhancing the sensitivity of peptide quantification for the targeted HCPs analysis
Host cell proteins (HCPs) are a major class of process-related impurities that accompany a recombinant biotherapeutic product during production. As their levels impact the potential toxicity and efficiency of the therapeutics, there are significant requirements for the quantitative measurement of HCPs across the entire development paradigm.
-
Technical note
Unlocking sensitivity for low-sample input quantitative proteomics using ZT Scan DIA on the ZenoTOF 8600 system
Unlock new possibilities in single cell proteomics analysis using ZT Scan DIA 2.0 on the ZenoTOF 8600 system to enable highly quantitative IDs from sub-nanogram sample amounts. In this work, ZT Scan DIA 2.0 boosts quantifiable protein groups and precursors up to 94% compared to traditional discrete window DIA methods, enabling deeper insights from minimal sample input.
-
Technical note
Glycoproteomics with the ZenoTOF 8600 system using electron activated dissociation (EAD)
Learn how the ZenoTOF 8600 system overcomes typical challenges in glycoproteomics with its improved sensitivity and tunable electron activated dissociation (EAD) capabilities. In this work, nanoflow LC separation and EAD fragmentation were used to identify N-glycopeptides enriched from depleted human plasma. Hybrid EAD-CID further demonstrated an increase in IDs by 54% relative to EAD alone.