Abstract
In this technical note, the ZenoTOF 7600 system was used to sensitively quantitate the ionizable lipid ALC-0315 and structurally elucidate its related impurities from lipid nanoparticles (LNPs) in plasma. Due to its inherent sensitivity, a triple quadrupole mass spectrometer (TQMS) is typically used for targeted bioanalytical quantitation. However, collision-induced dissociation (CID)-based lipid fragmentation is insufficient for their complete structural characterization. The ZenoTOF 7600 system is capable of sensitive, targeted quantitation and can utilize a complimentary fragmentation mode (electron-activated dissociation; EAD), which is effective for confident metabolic identification (MetID) (1-3) and the structural elucidation of lipids (4-9), thereby adding unique value for analytical versatility.
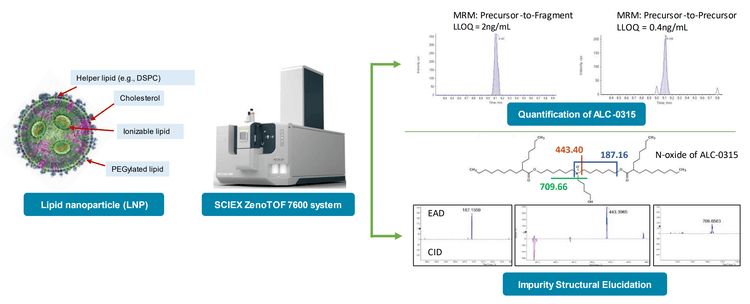
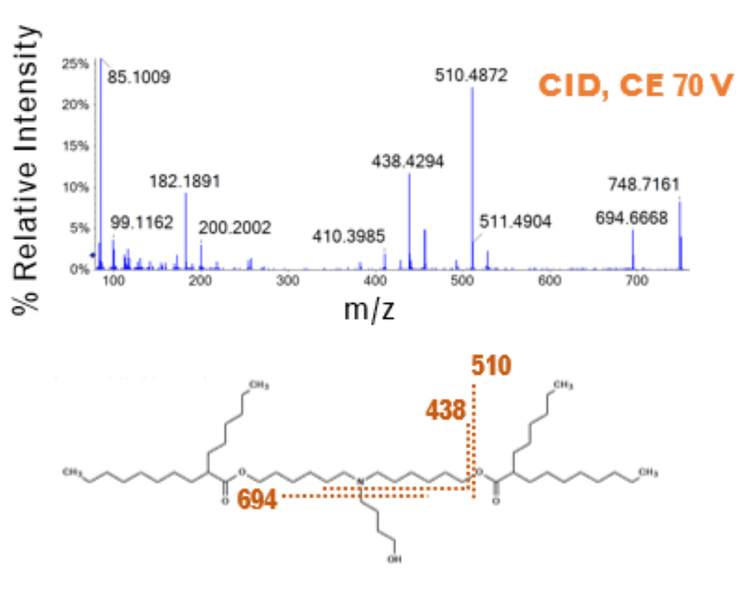
To demonstrate the quantitative performance of the ZenoTOF 7600 system for bioanalysis of ALC-0315, quantitation curves with lower limits of quantitation (LLOQs) were determined using either a precursor-to-fragment or precursor-to-precursor high-resolution MRM (MRMHR), and the amount of ALC-0315 from formulated LNPs diluted into plasma was then estimated. Impurities related to ALC-0315 at relative abundances as low as 0.02% were observed from the LNPs and some of the structures of these impurities were elucidated using EAD (Figure 1).
Key features of ionizable lipid analysis on the ZenoTOF 7600 system
- Through MRMHR acquisition, the ZenoTOF 7600 system can detect and quantitate ionizable lipids at sub ng/mL levels from formulated LNPs
- The simultaneous acquisition of full product ion spectra during quantitative analysis allows verification of other components within LNPs, such as the PEGylated lipid and structures related to the ionizable lipid
- EAD coupled with the Zeno trap allows for confident structural elucidation of low-level impurities associated with the ionizable lipid, demonstrating its usefulness in supporting MetID studies
Introduction
LNPs have emerged as safe and effective delivery vehicles for oligonucleotide-based therapeutics such as the siRNA-based Onpattro as well as messenger RNA (mRNA)-based vaccines such as Spikevax and Comirnaty (10). This has spawned investment into new precision genetic medicines by designing novel lipids to fine-tune properties like targeted delivery, biodegradation, and endosomal escape. LNPs typically comprise ionizable lipids, helper lipids such as distearoylphosphatidylcholine (DSPC), cholesterol, and PEGylated lipids (Figure 1). For example, the LNPs used in the Pfizer/BioNTech COVID-19 vaccine contained ALC-0315, DSPC, cholesterol, and PEG2000 conjugated lipid.
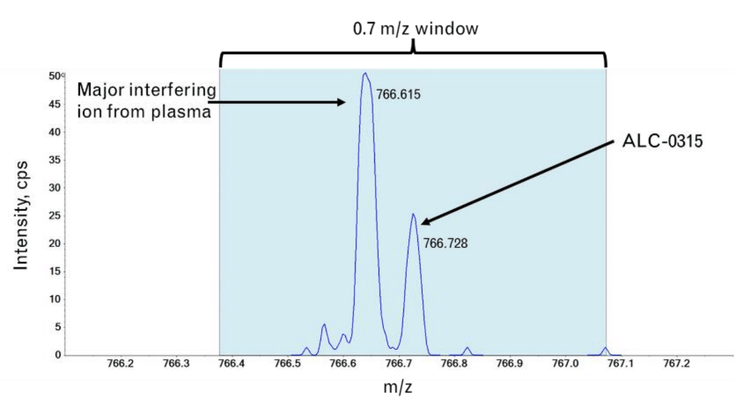
With the administration of novel exogenous materials, like the ionizable lipids used in LNPs, it is necessary to track their biodistribution and metabolism products to assess their safety and efficacy profiles. TQMSs are used for quantification studies to support bioanalytical assays using an MRM approach. However, some lipids may not fragment well by traditional CID fragmentation techniques nor produce diagnostic product ions to distinguish them from other interfering ions within biological matrices. In addition, it can be challenging to identify the structures of compounds formed from degradation or metabolism using a CID-based fragmentation alone. It has been shown that EAD-based fragmentation can produce information-rich spectra that can confidently elucidate structures of ionizable lipids and pinpoint sites of modifications within their related impurity structures (11-13).
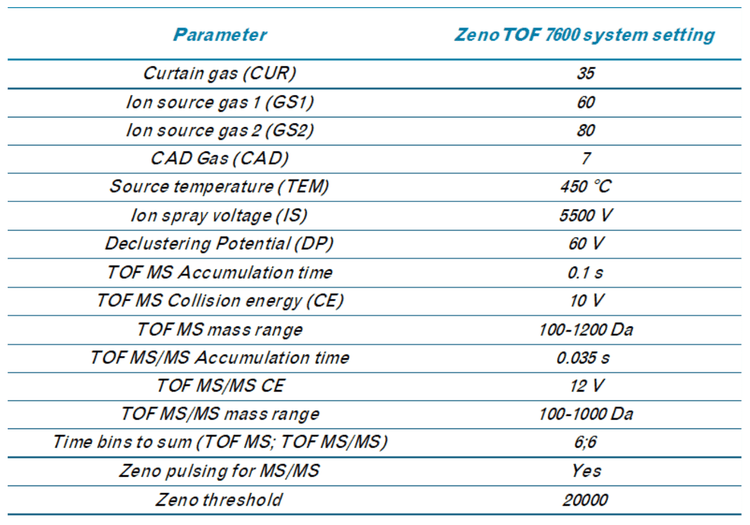
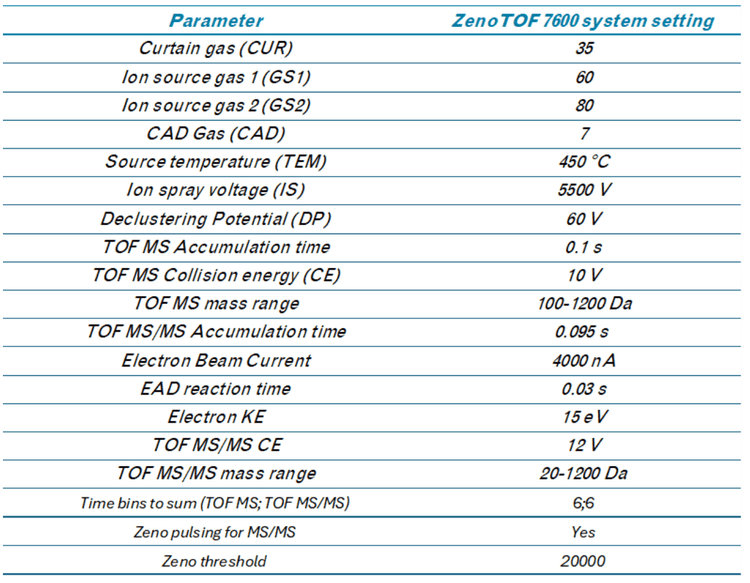
In this study, we assessed the ability of the ZenoTOF 7600 system to quantify ALC-0315 in plasma. This instrument includes a Zeno trap device that enhances MS/MS sensitivity to drive quantitative sensitivity. In addition, the instrument can provide complementary fragmentation via EAD to aid in structural characterization. Both aspects of the instrument were used to quantify and provide in-depth characterization of impurities related to ALC-0315 to demonstrate its utility in supporting LNP metabolism studies.
Materials and methods
Materials: ALC-0315 was purchased from Broadpharm (Cat #BP-25498) or Caymen Chemical (Cat #34337). DSPC (Cat #850365), cholesterol (CAT # 700100), and 1,2-dimyristoyl-sn-glycero-3- phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (ammonium salt) (PEG2000-PE) (CAT# 880150) were from Avanti Polar Lipids. The lipid components were diluted in EtOH before formulation. The nucleic acids, gWiz-GFP plasmid DNA (pDNA), and DasherGFP mRNA were from Aldevron. The nucleic acids were prepared in citrate-buffered saline at pH 4.25. All solvents were LCMS grade and obtained from Burdick and Jackson. Human control plasma was from SCIEX.
LNP Formulation: To produce LNPs, lipids were mixed at a molar ratio of ALC-0315Cholesterol: PEG2000-lipid 46.3:9.4:42.7:1.6. LNPs were formulated with a NanoAssemblr Ignite microfluidic device from Cytiva using a 10 mL/min flow rate and a 3:1 mixing ratio of aqueous citrate buffer, which contains the nucleic acid, to EtOH, which contains the lipid mixture. Following formulation, LNPs were diluted with phosphate-buffered saline and a buffer exchange was performed using 100kDa molecular weight cut-off ultracentrifugal filters.
Sample preparation: ALC-0315 and LNPs were diluted into plasma. Samples were extracted by protein precipitation by adding 100% ACN, followed by benchtop centrifugation for 10 minutes. Supernatants were collected, and 2 mL were injected into the LC column for LC-MS/MS analyses.
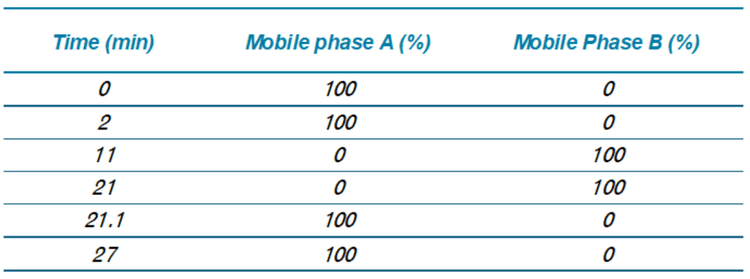
Chromatography: Extracts were analyzed on a ZenoTOF 7600 system using an Exion UHPLC. Samples were separated with a Peptide BEH C18 column (150 x 2.1 mm; 1.7 µm, 130 A, Waters, P/N:186003687). Chromatographic gradient details are shown in Table 1. The autosampler and column oven temperatures were 15 °C and 70 °C, respectively. The mobile phase flow rate was 0.5 mL/min, and the total run time was 27 min. Mobile phases A and B consisted of 10 mM ammonium acetate in water/MeOH/ACN (15/30/55; v/v) and 10 mM ammonium acetate in ACN/MeOH (60/40; v/v), respectively
After establishing the quantitative methodology for ALC-0315, we applied this to formulated LNPs. LNPs were diluted in methanol and subjected to LC-MS analysis for initial assessment. All components of the LNPs were observed, including ALC-0315 and the PEGylated lipid (Figure 6). This indicated that a bioanalytical method could be developed for other exogenous materials like the PEGylated lipid in addition to the ionizable lipid using this methodology.
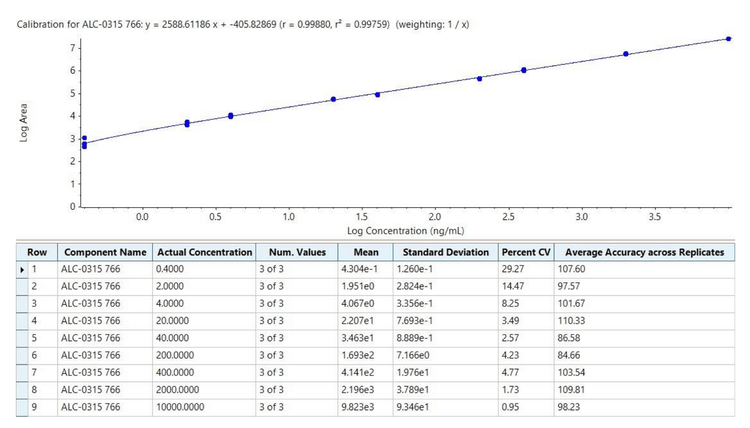
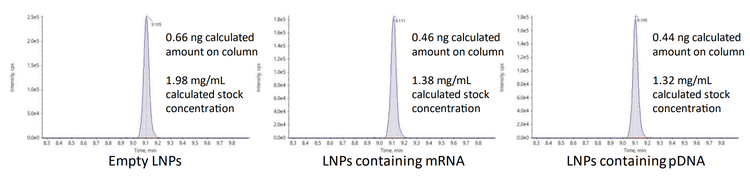
The amounts of ALC-0315 from three different preparations of LNPs diluted into plasma were measured to assess the quantitative performance of the ZenoTOF 7600 system. The LNPs were initially formulated with mRNA, pDNA, or they were empty. The peak areas of the ALC-0315 detected from these LNP preparations diluted 1:6000 are shown in Figure 5. The concentrations of ALC-0315 from 3 LNP preparations were calculated as 0.33, 0.23, and 0.22 mg/mL, based on the external calibration curve from Figure 4. These measurements estimated the original stock concentrations to range from 1.32 to 1.98 mg/mL by extrapolation. This is consistent with the amount of ALC-0315 initially used in the LNP formulation, allowing for some moderate sample loss, where 20-50% lipid loss from filtration (e.g., dialysis) is typical (14-15). A dilution of 1:6000 was 1000x higher than the LLOQ, demonstrating the sensitivity of the ZenoTOF 7600 system. Overall, this method is suitable for measuring trace amounts of material for thorough and sensitive biodistribution assessments.
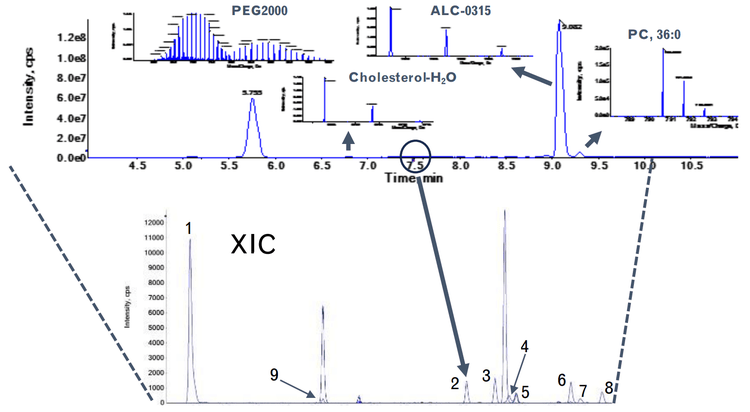
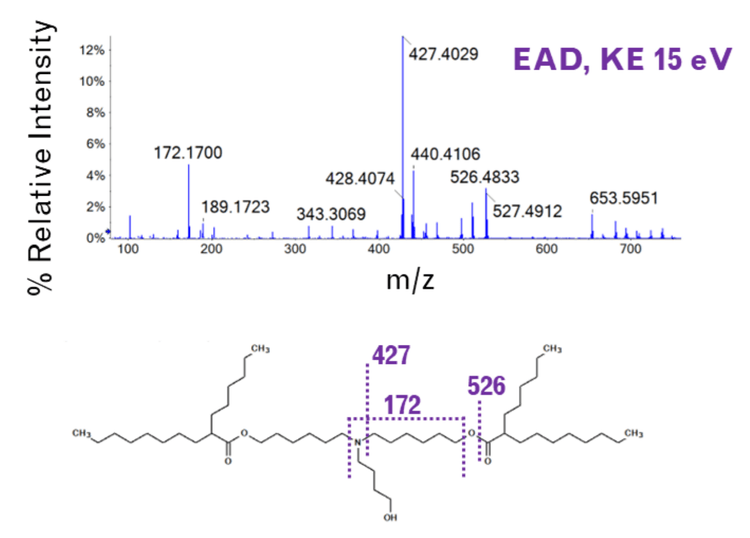
EIEIO-based fragmentation was used on selected impurities of ALC-0315 after LNP dilution into plasma. Unique fragments of m/z 187.156, 443.397, and 709.656 were observed for an oxidative impurity of m/z 781.716 by EIEIO, which were not seen by CID. (Figure 1, lower right). These fragments were used to pinpoint oxygen incorporation to a specific, confined region surrounding the tertiary amine within the structure of ALC-0315, strongly suggesting that this impurity was the N-oxide of ALC-0315.
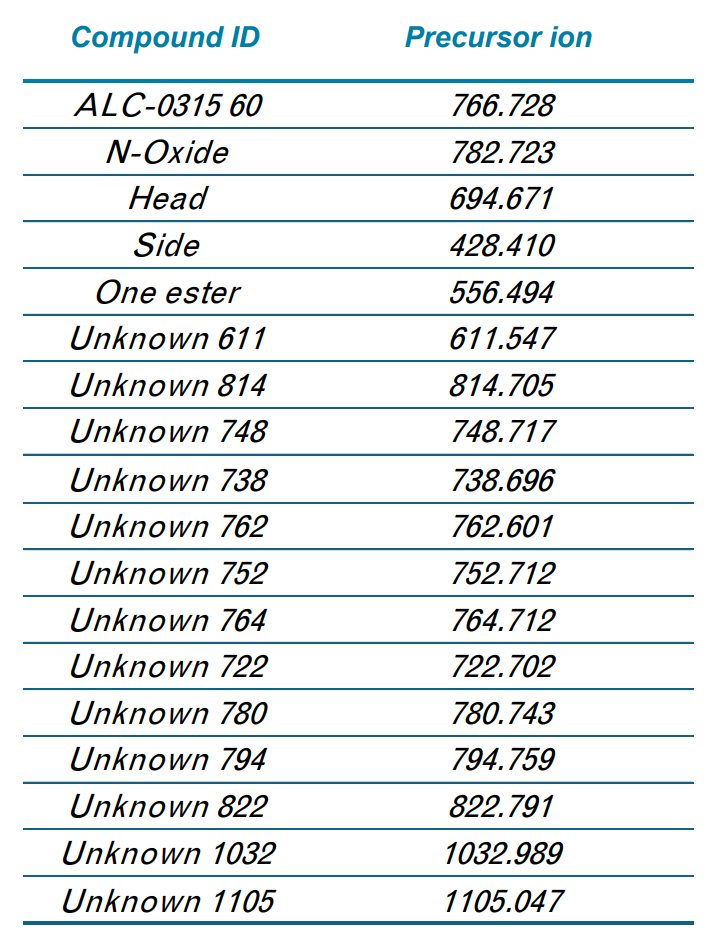
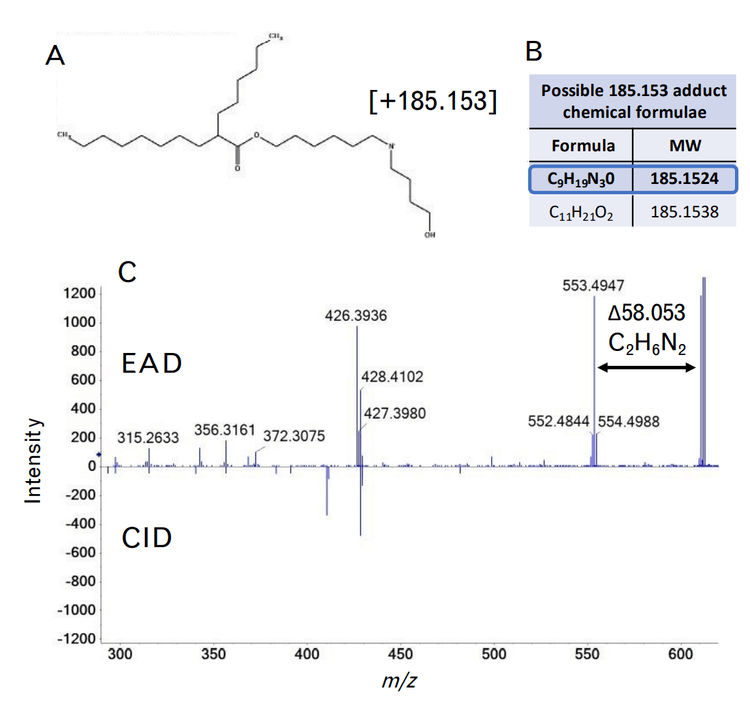
A more complex unknown impurity at m/z 610.540 was also observed. This molecular weight is consistent with the loss of one of the hexane-6,1-diyl bis(2-hexyldecanoate) (C22H44O2) chains, and the addition of an unknown covalent addition with a molecular weight of 185.153 (Figure 8A). This molecular weight could be explained by two different chemical formulae based solely on MS accurate mass – C9H19N3O or C11H21O2, as shown in Figure 8B. Fragmentation by EAD resulted in a unique fragment of m/z 553.494 not seen by CID, which was 58.053 mass units less than the precursor molecular weight (Figure 8C). This loss could only be explained by a nitrogen-containing chemical formula of C2H6N2 due to accurate mass measurement and the mass defect of molecular nitrogen. In addition, fragment ions matching losses relating to the chemical formulae of C8H19N2 and C9H21N2 were also observed in the EAD fragment ion spectrum. As a result, the adducted chemical formula was narrowed down to C9H19N3O. Although it is difficult to derive the exact structure of this adduct, these data show that EAD provides valuable information that is not attainable by accurate mass measurements or by CID alone. This information could help corroborate the elucidation of other unknown lipid-adducted species from oligonucleotide or protein modifications.
Conclusion
- The sensitivity of the ZenoTOF 7600 system enables the low-level detection and quantitation of lipids related to LNPs in plasma to support quantitative bioanalysis in complex matrix
- Fragmentation of ionizable lipids and their related impurities using EAD results in information-rich product ion spectra even at low abundance after dilution into plasma
- Unique fragment ions of impurities related to ALC-0315 from EAD can be used to confidently identify these analytes and pinpoint sites of modification, demonstrating that this technique can support and benefit metabolism studies
- Sensitive quantitation paired with detailed structural characterization shows the versatility of the ZenoTOF 7600 system to support multiple workflows within DMPK
References
- Che P, Davidson JT, Kool J, Kohler I. Electron activated dissociation – a complimentary fragmentation technique to collision-induced dissociation for metabolite identification of synthetic cathinone positional isomers. Anal Chim Acta. 2023; Dec 1;1283 1-12. doi: 10.1016/j.aca.2023.341962
- Calabrese V, Brunet TA, Degli-Esposti D, Chaumot A, Geffard O, Salvador A, Clement Y, Ayciriex S. Electron-activated dissociation (EAD) for the complementary annotation of metabolites and lipids through data-dependent acquisition analysis and feature-based molecular networking, applied to the sentinel amphipod Gammarus fossarum. Anal Bioanal Chem. 2024; May;416(12):2893- 2911. doi 10.1007/s00216-024-05232-w
- He Y, Hou P, Long Z, Zheng Y, Tang C, Jones E, Diao X, Zhu M. Application of electro-activated dissociation fragmentation technique to identifying glucuronidation and oxidative metabolism sites of vepdegestrant by liquid chromatography-high resolution mass spectrometry. Drug Metab Dispos. 2024; Jun 17;52(7):634-643. doi 10.1124/dmd.124.001661
- Baba T, Campbell JL, Le Blanc JCY, Baker PRS. In-depth sphingomyelin characterization using electron impact excitation of ions from organics and mass spectrometry. J Lipid Res. 2016; May;57(5):858-67. doi 10.1194/jlr.M067199
- Baba T, Campbell JL, Le Blanc JCY, Baker PRS. Distinguishing cis and trans isomers in intact complex lipids using electron impact excitation of ions from organics mass spectrometry. Anal Chem. 2017; Jul 18;89(14):7307-7315. doi: 10.1021/acs.analchem.6b04734
- Baba T, Campbell JL, Le Blanc JCY, Baker PRS. Structural identification of eicosanoids with ring structures using differential mobility spectrometry-electron impact excitation of ions from organics mass spectrometry. J Am Soc Mass Spectrom. 2023; Jan 4:34(1);75-81. doi: 10.1021/jasms.2c00256
- Baba T, Campbell JL, Le Blanc JCY, Baker PRS. Structural identification of triacylglycerol isomers using electron impact excitation of ions from organics (EIEIO). J Lipid Res. 2016; Nov;57(11);2015-2027. doi: 10.1194/jlr.M070177
- Pearson M, Hunter C, Baba T. Complete structural elucidation of lipids in a single experiment using electron activated dissociation (EAD). SCIEX technical note, RUO-MKT-02-13050-B
- Jinmei C, Dandan S, Zhimin L, Lihai G. Systematic determination of lipid structure using electron activated dissociation (EAD). SCIEX technical note, RUO-MKT-02- 14182-A
- Chaudhary N, Weissman D, Whitehead KA. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discov 2021; Nov;20(11):817– 838. doi: 10.1038/s41573-021-00283-5
- Crowe A, Jain N, Higgins R, Proos R, Stone M, Pohl K. Distinguishing oxidative impurities from ionizable lipids used in LNP formulations using electron activated dissociation. SCIEX technical note, RUO-MKT-02-14983-A
- Yang Z, Baker P, Mollah S, Proos R, Le Huray J. Structural characterization of the cationic lipid nanoparticle component, ALC-0315, and its impurities using electron-activated dissociation (EAD)-based MS/MS fragmentation. SCIEX technical note, RUO-MKT-26966-A
- Yang Z, Mollah S, Kaluarachchi H, Jin W, Al-Dulaymi M. Automated characterization of the lipid nanoparticle ionizable lipid, DODAP, and its degradants using Molecule Profiler software. SCIEX technical note, RUO-MKT-26226-A
- De Peña AC, Zimmer D, Gutterman-Johns E, Chen NM, Tripathi A, Bailey-Hytholt CM. Electrophoretic microfluidic characterization of mRNA- and pDNA-loaded lipid nanoparticles. ACS Appl Mater Interfaces. 2024; May 29;16(21):26984-26997. doi: 10.1021/acsami.4c00208
- Mousli Y, Brachet M, Chain JL, Ferey L. A rapid and quantitative reversed-phase HPLC-DAD/ELSD method for lipids involved in nanoparticle formulations. J Pharm Biomed Anal. 2022; Oct 25:220:115011. doi: 10.1016/j.pba.2022.115011
- https://downloads.regulations.gov/CDC-2021-0034- 1148/attachment_1.pdf