Abstract
Recycled tire products pose risk to both the environment and human health due to the recently identified quinone transformation product of N-(1,3-dimethylbutyl)-N′-phenyl-p-phenylenediamine (6PPD). Due to the wide variety of compounds present in tire derived samples and the presence of thousands of unknown features, comprehensive screening methods and comprehensive data collection methods detection are required. Using the SWATH acquisition workflow on the X500B QTOF system and SCIEX OS software, 18 compounds were discovered in the tire derived samples, including 6PPD-quinone, which has been shown to be extremely toxic to certain salmon species.
Introduction
Recycled tire products pose risk to both the environment and human health.1,2 A recent study by Tian et al. (2021) identified the presence of a quinone transformation product of N-(1,3-dimethylbutyl)-N′-phenyl-p-phenylenediamine (6PPD), as a potential source of acute mortality in U.S. Pacific Northwest Coho salmon (Oncorhynchus kisutch).2 6PPD is used as a tire rubber antioxidant and it, along with its transformation product 6PPD-quinone, were found in roadway runoff and stormwater samples at toxic concentrations.2
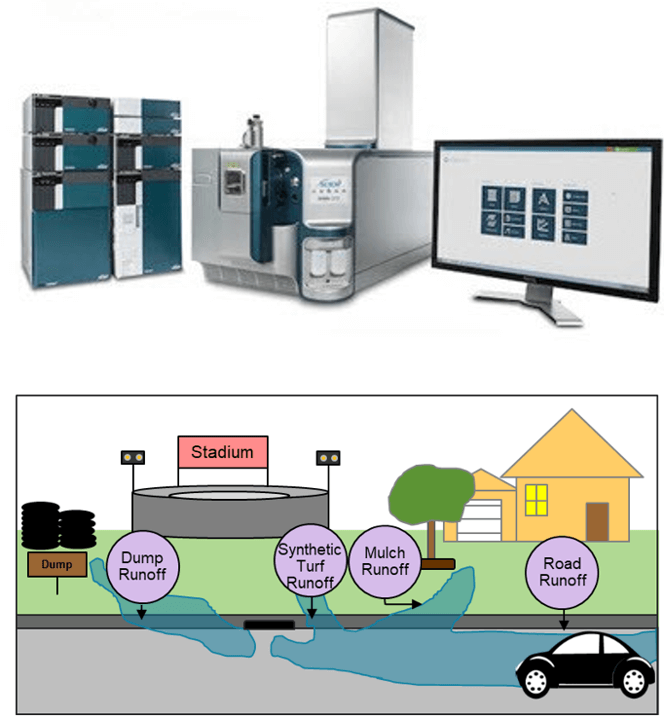
While roadway runoff is one pathway for tire related compounds to enter the environment, there are many other ways these compounds can be introduced. Recycled tire products have been used for a variety of purposes including landscaping, rubberized asphalt, and as a synthetic alternative for turf fields. These synthetic turf fields have been installed in the United States since the 1960s and currently, there are 12,000-13,000 synthetic turf fields nationally, with 1,200 – 1,500 new fields being installed each year. According to the United States Environmental Protection Agency (US EPA), 6PPD was observed in nontargeted analysis of both recycling plant and synthetic turf field samples with relatively high response area counts, along with many other compounds that were tentatively identified or not identified.
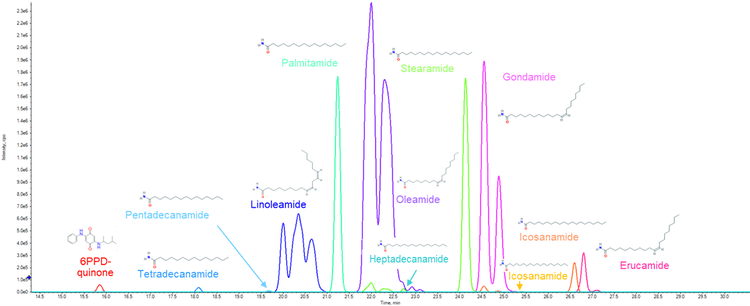
Due to the wide variety of compounds present in tire derived samples (Figure 1) and the presence of thousands of unknown features, comprehensive screening methods and comprehensive data collection methods detection are required. This study uses the X500B QTOF system and a data independent acquisition approach (SWATH acquisition) to acquire high-quality, high-resolution MS/MS data to screen for both known-unknowns and perform nontargeted analysis. In the tire derived sample, 17 amide compounds were identified in addition to 6PPD-quinone. Of these 17, several compounds were not found in mass spectral databases or in the literature, as they are likely impurities created in the tire production process.
Key features of the X500B QTOF system and SCIEX OS software
- SWATH acquisition on the X500B QTOF system acquires MS/MS spectra on all detectable compounds, providing a comprehensive fragmentation map for identification and quantification
- Advanced processing features including custom columns in SCIEX OS software 2.1.6 allow for “on the fly” blank filtering and feature prioritization
Methods
Sample preparation: Tire derived samples were collected in 10 mL PTFE sampling bottles and passed through a 0.2 μm PTFE syringe filter before analysis.
Chromatography: LC separation was achieved using the SCIEX ExionLC AD system and a Phenomenex Luna Omega Polar C18 column (3 µm, 3.0 x 100 mm). A 45-minute gradient of water and methanol with 0.1% formic acid buffer was used, with a flow rate of 0.6 mL/min and column temperature of 40 ̊C.
Mass spectrometry: Mass spectrometry was performed using the X500B QTOF system, with electrospray ionization (ESI) in positive mode. TOF MS and SWATH acquisition experiments were performed. Data acquisition was performed using SCIEX OS software 2.1.6.
Data processing: Processing of MS data was performed using SCIEX OS software 2.1.6 in which blank filtering, library matching, and formula finding were performed. SIRIUS was also used for database searching.3 SIRIUS uses isotope pattern analysis and fragmentation trees with structural elucidation to provide a likely parent formulas from large databases like PubChem.3 This combined approach leads to more accurate predictions and identification rates of more than 70% on challenging datasets.3
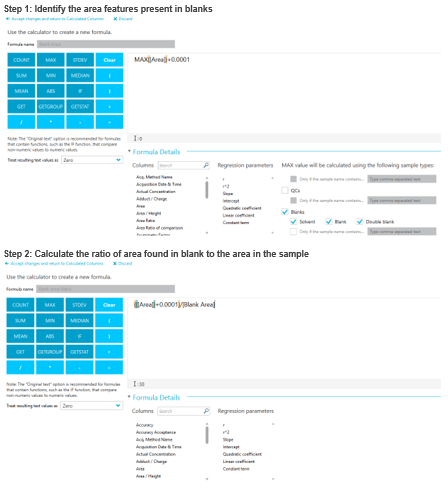
Blank filtering using SCIEX OS software and custom columns
Blank filtering is a crucial step in nontarget analysis to ensure time is not spent on features present in instrument and extraction blanks. In SCIEX OS software 2.1.6, this can be done using calculated columns and custom filtering (Figure 2). During this same step, gap filling can be applied to ensure errors that would occur due to attempting to divide by zero can be eliminated by adding a small area (0.0001) to each feature. In this study, a blank area threshold of 10 was used to determine which features were present only in the tire derived samples. Using this approach, the number of found features decreased from 174 to 34, thus providing 34 features unique to the tire derived samples to be identified.
Suspect screening approach
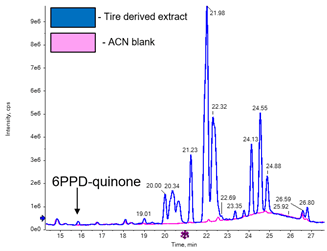
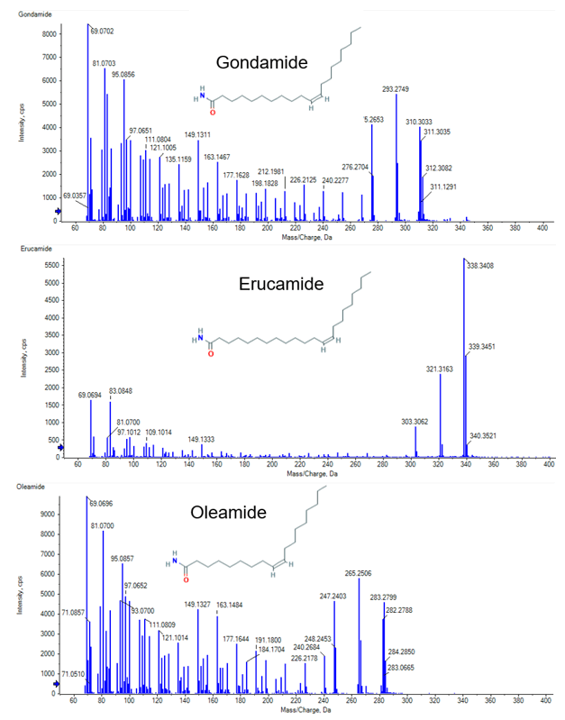
A suspect list was compiled from the EPA 2019 tire crumb rubber characterization report, and the newly identified 6PPD-quinone was also added.1,2 This list contained 111 positively ionized compounds and from this list, two compounds were identified and later confirmed with MS/MS library spectra: 6PPD-quinone (Figure 3) and oleamide. Oleamide is a common processing additive used in manufacturing tires and other molded and extruded products.
Nontarget screening approach
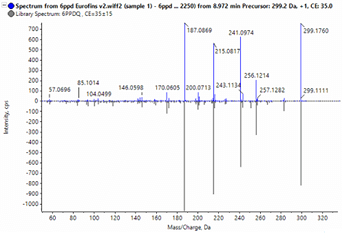
The remaining 32 features were processed using a nontarget workflow. The feature list was first sorted by the ratio of the compound in the blank and the area in the samples. This approach was taken to prioritize the features most unique to the samples. From this, formula finding, adduct identification, and NIST ’17 MS/MS Library searching were performed using Analytics in SCIEX OS software. Two additional compounds were identified from the NIST ’17 MS/MS library search: stearamide (C18H37NO) and palmitamide (C16H34NO), both common additives in tire production. The results from the formula finding in SCIEX OS software were compared to the formula results from SIRIUS.3 Matching top formulas were then prioritized and 2 features with a similar formula to oleamide, differencing by only the number of -CH2- units, were identified, thereby leading to the discovery of erucamide (C22H43NO), another additive used in tires and molded rubber products, and gondamide (C20H39NO) (Figure 5).
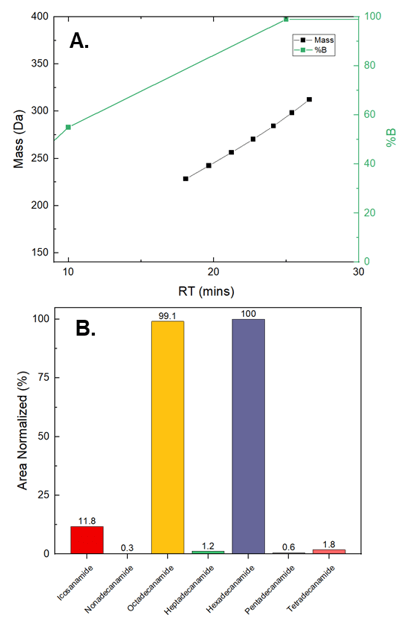
To further screen the unknown features, a suspect list was built that contained all the theoretically possible homologues of gondamide and stearamide differing only by -CH2- units. An example of this approach is shown in Figure 4. Here, 7 features are shown to differ only by the number of -CH2- units and have the same retention time shift between homologs. Using this approach, 11 addition features were identified and were confirmed using diagnostic ions, retention time shift, and mass accuracy. Of these 11, several had the same accurate mass and fragmentation, but existed at different retention times due to the existence of cis and trans isomers (Figure 6).
Conclusions
As human populations continue to grow and continue to encroach upon aquatic environments, the impact of chemicals derived from tires will continue to increase. These complex mixtures require high resolution LC-MS/MS workflows and advanced software tools for characterization. Using the SWATH acquisition workflow on the X500B QTOF system and SCIEX OS software, 18 compounds were discovered in the tire derived samples, including 6PPD-quinone, which has been shown to be extremely toxic to certain salmon species.2 In addition to 6PPD-quinone, multiple other compounds known to be used during tire production were discovered, while several others are likely impurities present in these known compound mixtures.
References
- United States Department of Environmental Protection. Synthetic Turf Field Tire Crumb Rubber Research Under the Federal Research Action Plan Final Report Part 1 – Tire Crumb Rubber Characterization Appendices; 2019; Vol. 2.
- Tian Z et al. (2021) Ubiquitous tire rubber–derived chemical induces acute mortality in Coho salmon. Science 371 (6525), 185–189.
- Dührkop K et al. (2019) SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 16 (4), 299–302.