Abstract
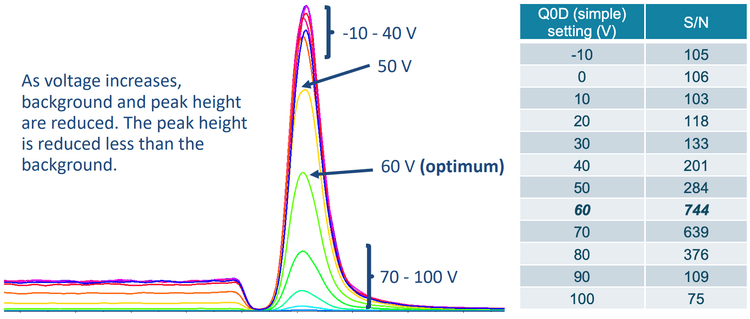
In this technical note, sensitive analysis of 13 nitrosamines in a metformin API was performed using the SCIEX 7500 system with limit of quantitation (LOQ) values as low as 0.01 ng/mL in solution. Q0D, a feature unique to the SCIEX 7500 system, was applied to improve the signal-to-noise (S/N) ratio of low molecular weight impurities, such as NDMA (Figure 1).
Introduction
Nitrosamines have been a concern in the pharma industry for several years due to their carcinogenic health effects. These concerns are increasing following the discovery of complex and API-specific nitrosamine impurities. A recent study indicated that up to 40% of all APIs might be susceptible to nitrosamine formation. 1 Therefore, improved analytical methodologies are needed to test and screen drug products and APIs for these impurities.
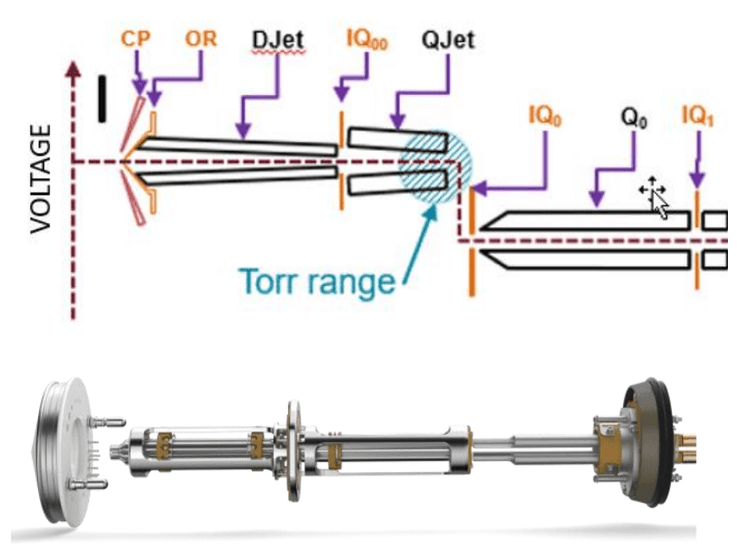
Key features of nitrosamine analysis using the SCIEX 7500 system
- Sub-ng/mL level quantitation: LOQs as low as 0.01 ng/mL were achieved for nitrosamine analysis on the SCIEX 7500 system when applying Q0D
- Ideal analytical performance: Accurate quantitative performance with %CV <8.70% achieved at the LOQ for each compound and across linear dynamic ranges (LDRs) spanning up to 3.5 orders of magnitude
- High sensitivity with small sample amount: Two sample preparations (50 and 5 mg/mL) were used to demonstrate that limits can be achieved even with low sample amounts
- Enhanced sensitivity unlocked: Improved front-end technology with the D Jet ion guide, OptiFlow Pro ion source and E Lens probe enhanced the generation, capture and transmission of ions2
- Streamlined data management: Fast, intuitive and integrated data acquisition and processing were performed using SCIEX OS software
Methods
Standard preparation: Nitrosamine standards were provided as a mixed solution at 1 µg/mL. This solution was diluted in water to cover concentrations ranging from 0.002 to 50 ng/mL.
Sample preparation: Samples of 5 or 50 mg of metformin API were weighed into individual Eppendorf tubes. Then, 1 mL of water was added to each tube before it was vortexed to mix and dissolve the metformin API.
Spiked sample preparation: Samples of 5 or 50 mg of metformin API were weighed into individual Eppendorf tubes. Then, 1 mL of a 0.1 ng/mL mixed standard was added to each tube before it was vortexed to mix and dissolve the metformin API.
Chromatography: Separation was performed using an ExionLC AD system and a Phenomenex biphenyl column (100 x 3 mm, 2.6 µm, 100 Å). Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in methanol. The flow rate was 0.75 mL/min and the column temperature was 40°C. The injection volume was 30 µL. Diverter settings were as follows: 0–1.9 min to waste, 1.9–10.5 min to MS and 10.5–16 min to waste. The gradient program used is described in Table 1.
Mass spectrometry: The SCIEX 7500 system was operated in positive ion mode using atmospheric pressure chemical ionization (APCI). Scheduled MRM acquisition was utilized for analysis. The MRM transitions and optimized collision energy (CE) values used are included in Table 2.
Data processing: SCIEX OS software was used for data acquisition and processing.
How Q0D enables ultra high levels of sensitivity
Sensitivity is critical for nitrosamine analysis due to the regulations provided by both the EMA and FDA. The most potent nitrosamines have a requirement limit of 26.5 ng/day. 3-4 This limit within a formulated product is dependent on the maximum daily dose (MDD) of the specific medication. The FDA requires a limit of 0.03 µg/g for medicines with a daily dosage <880 mg. This equates to a concentration in a solution of 0.15 ng/mL (5 mg sample preparation) or 1.5 ng/mL (50 mg sample preparation). The EMA states that skip testing can be performed if the LOQ of the method is ≤30% of the specified limit and/or omission of the specification can be justified if the LOQ is ≤10% of the limit. These LOQ values equate to 0.045 or 0.015 ng/mL for a 5 mg sample preparation or 0.45 or 0.15 ng/mL for a 50 mg sample preparation, assuming a daily dosage of <880 mg. Metformin can have a higher MDD than 880 mg. Here, analysis is performed on the API and not the final drug product.
Q0D on the SCIEX 7500 system was employed to enhance quantitative sensitivity and meet regulatory limits. In simple mode, Q0D is a voltage that is applied between the QJet ion guide and the IQ0 lens. Simple mode is used to break up ion clusters and reduce the noise level to increase S/N values (Figures 1 and 2).
Enhanced Q0D is stronger than simple Q0D and is used to fragment the compound. This approach uses 2 fragmentations to perform a pseudo MS3 experiment. Further information about Q0D can be found in reference 5.
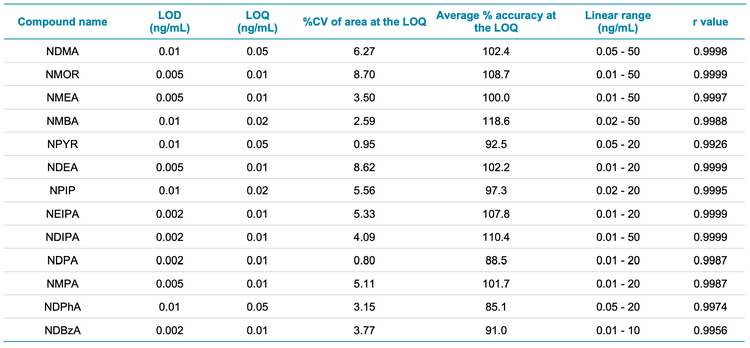
Q0D was optimized for the compounds analyzed. The LOQ and LOD values achieved are shown in Table 3.
Quantitative sensitivity
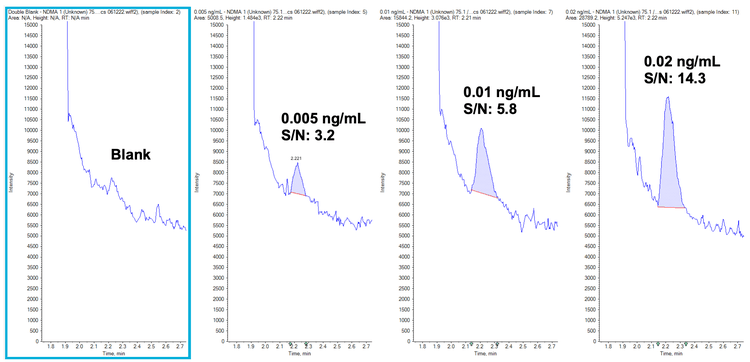
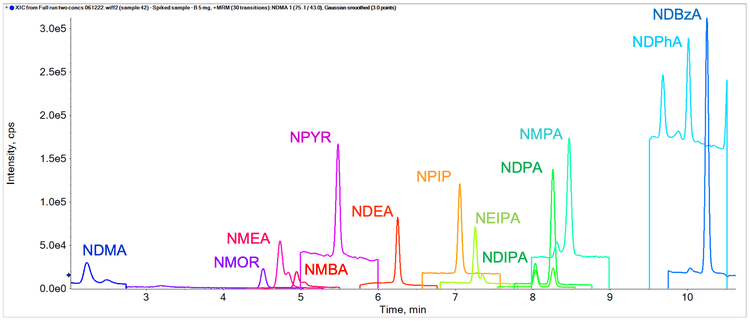
The precision, accuracy and linear range of the assay were assessed and passed the specifications for trace level analysis. These criteria included %CV values <10% at the LOQ, % accuracy between 70% and 130% across all levels of the calibration curve and r values >0.99. Table 3 shows the precision, accuracy and linear range results achieved. The sensitivity of the analysis for NDMA is highlighted in Figure 3 and Figure 4 shows extracted ion chromatograms (XICs) of all nitrosamines at their respective LOQs.
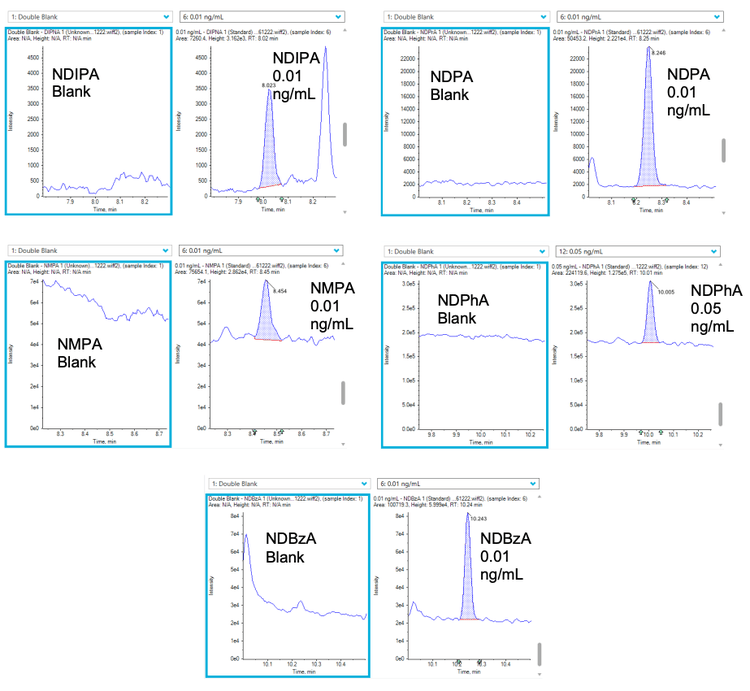
Sample preparation for spiking experiments
The sensitivity of the SCIEX 7500 system allows low sample concentrations to be analyzed at desired sensitivity levels. For this analysis, 2 sample preparations were employed (50 mg/mL and 5 mg/mL). Duplicate sample preparations were spiked at 0.1 ng/mL and each was injected in triplicate to assess accuracy and precision. Figure 5 shows the overlaid XICs of all spiked nitrosamines.
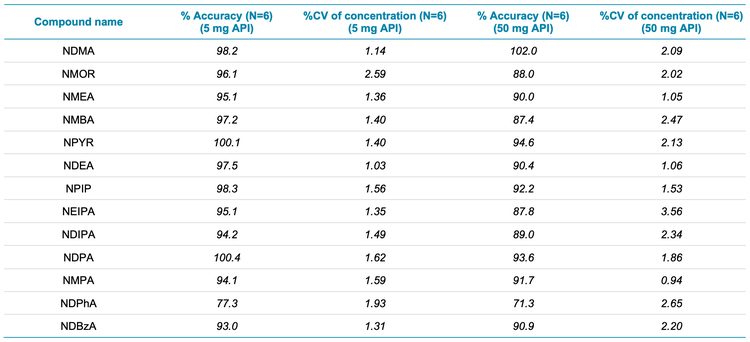
Table 4 compares the accuracy and precision values achieved for both sample preparations, based on 6 injections of each preparation. All compounds analyzed provided % accuracy values between 70% and 130% and precision values <10 %CV. These results indicate that the method is capable of precise and accurate quantitation of nitrosamines present at trace levels.
Conclusion
- A sensitive method for nitrosamine quantitation was developed using Q0D on the SCIEX 7500 system to achieve LOQ values as low as 0.01 ng/mL
- The method demonstrated accurate and highly reproducible (%CV <8.70%) results at the LOQ of each compound. Accuracy, precision and linearity passed specifications for trace level analysis
- Sensitive analysis of small sample amounts was achieved for 2 sample preparations at 50 mg/mL and 5 mg/mL, with spiking performed at 0.002 and 0.02 µg/g respectively
- The precision and accuracy results for the sample spiked at 0.1 ng/mL met specifications, with % accuracy between 70% and 130% and %CV <10%
- High sensitivity was achieved on the SCIEX 7500 system with improved front-end technology for better ion generation, capture and transmission
- A single platform was used for streamlined data acquisition, processing and management with SCIEX OS software
References
- The Landscape of Potential Small and Drug Substance Related Nitrosamines in Pharmaceuticals, Joerg Schlingemann et al, Journal of pharmaceutical sciences, Global health, Volume 112, Issue 5, November 2022
- Enabling new levels of quantification. SCIEX technical note, RUO-MKT-02-11886-A.
- Questions and answers for marketing authorization holders/applicants on the CHMP Opinion for the Article 5(3) of Regulation (EC) No 726/2004 referral on nitrosamine impurities in human medicinal products, EMA, March 2023
- Control of nitrosamine impurities in human drugs, FDA, February 2021
- Two approaches for MRM3 data acquisition using the SCIEX 7500 system. SCIEX technical note, RUO-MKT-02- 14214-A.


