Powerful Processing and Interpretation for MetID
MetabolitePilot™ Software 2.0 is the industry leading metabolite identification software from SCIEX. It gives you the flexibility and power to analyze small molecule metabolites and the most complex large molecule catabolites – faster and more efficiently than ever before. It is designed specifically to enhance SCIEX TripleTOF® 5600+ and 6600 systems, and is compatible with the data produced by Analyst®TF Software.

Flexible
Save time with this all-in-one software that has the flexibility to handle small molecule metabolite and large molecule catabolite analysis. It delivers advanced identification, interpretation, visualization and correlation of a wide variety of molecule types.
Powerful
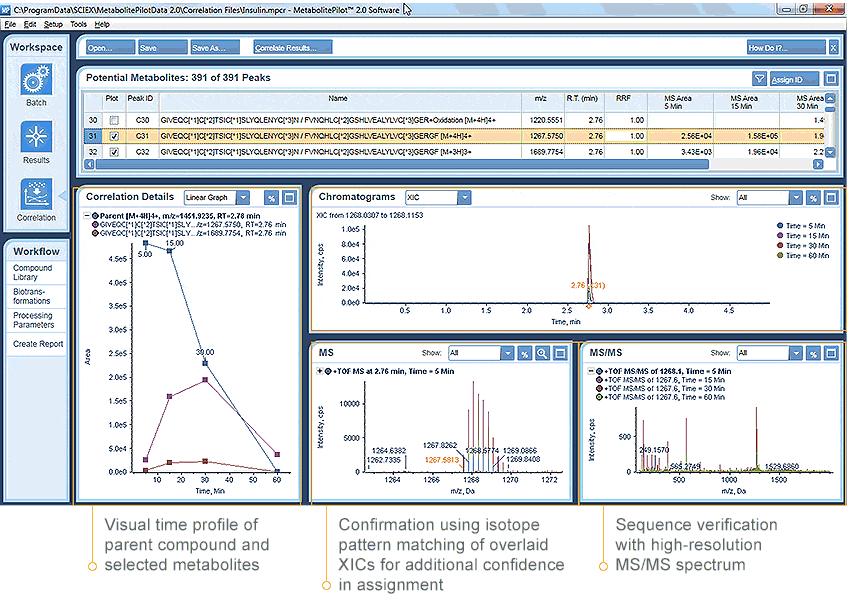
You can quickly and easily correlate metabolites and catabolites across multiple samples and automatically generate plots, overlay chromatograms or visualize MS and MS/MS data, all within a single simplified user interface.
Efficient
Metabolite and catabolite identification is now even faster, with batch data import and automated method generation for up to 200 samples at a time, giving you more time for decision-making and to focus on results.
Comprehensive
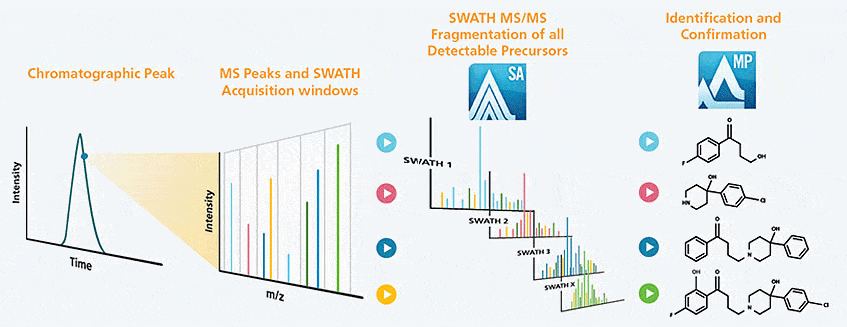
You have the ultimate confidence that you are seeing all detectable metabolites in your sample, with a single injection analysis using SWATH® Acquisition and MetabolitePilot Software data processing and interpretation.
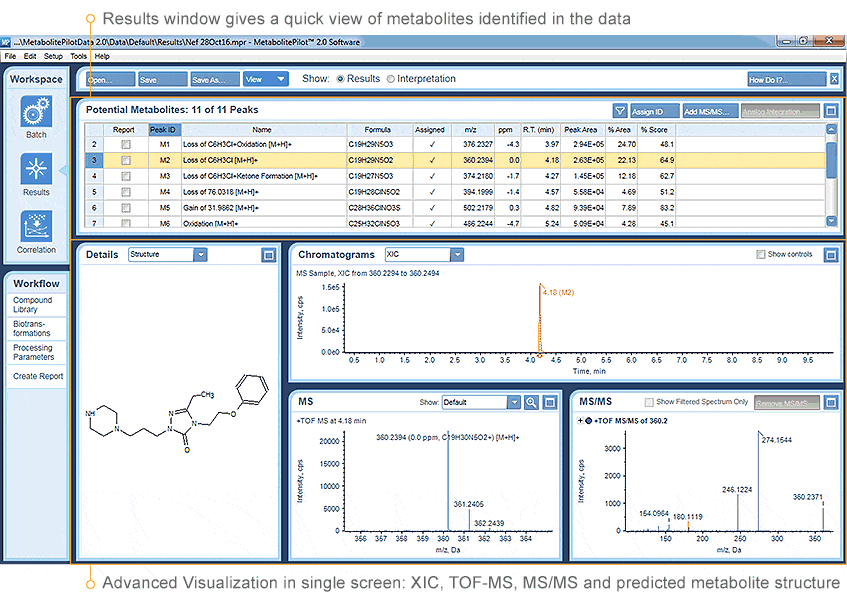
Confident, Automated Identification and Confirmation
MetabolitePilot Software quickly and accurately interprets and correlates metabolite structures and profiles. You will always see your data in context, based on your data acquisition strategy and within the confidence range that you define.
Using multiple powerful processing algorithms, MetabolitePilot Software simplifies the complexity of MetID for both small and large molecule analysis. It gives you the tools to perform both non-targeted and targeted processing in parallel to quickly identify metabolic and catabolic hotspots. The automated structure generation feature enables you to efficiently detect metabolites, minimizing data review by ranking proposed structures/sequences based on high-quality accurate MS/MS.
Accelerate Your Workflows with Confident Results
From structural elucidation and cross correlation to kinetic trends and reporting, MetabolitePilot Software enhances the advanced acquisition workflows of the SCIEX TripleTOF systems for definitive metabolite detection. You can use the same comprehensive SWATH Acquisition workflow for both small and large molecule metabolism studies, paired with the MetabolitePilot software you will significantly shorten your time to answers and have the greatest confidence in your results.
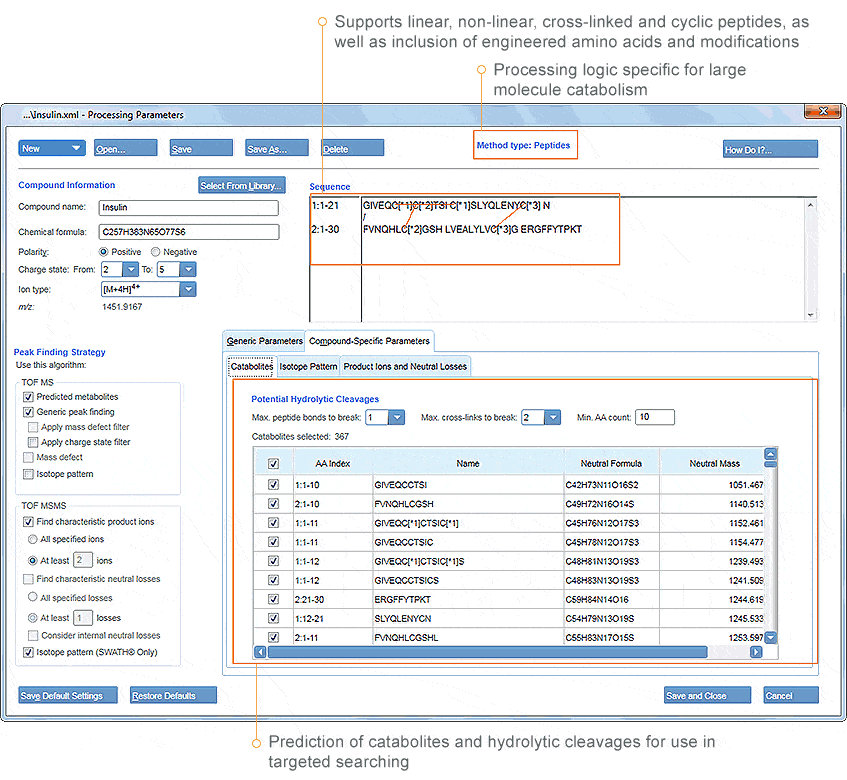
Advanced Processing for Biotherapeutic Catabolism
With MetabolitePilot Software, you can employ the same automated processing and assignment for large molecule catabolites as you can for drug metabolites. This means you can expect fast, comprehensive and accurate peptide catabolism and biotransformation identifications, enabling you to find liabilities and soft spots in the molecule, which may keep it from being biologically active and safe, in vivo.
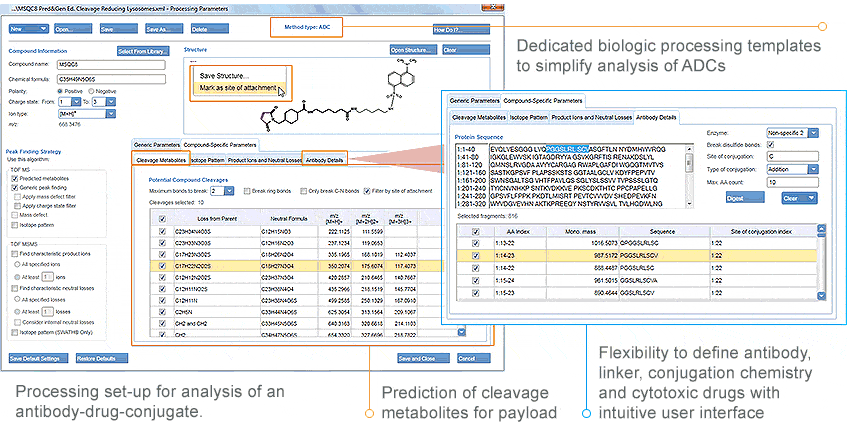
Antibody Drug Conjugates Made Easier
Although ADCs are inherently complex, MetabolitePilot Software helps to make them easier to analyze. By harnessing the intrinsic intelligence of the software, you can simplify ACD metabolism with dedicated processing templates and advanced logic to help you get to answers faster than ever before. You will be confident in your ADC assignments with the new interactive results view, where you can confirm metabolite, catabolite and biotransformation structures.
See More Than You Thought Possible
SCIEX proprietary SWATH Acquisition captures 100% high-resolution MS/MS spectra, so you can be certain that you are catching all detectable metabolites and catabolites in your sample. Moreover, the high-quality MS/MS data from SWATH Acquisition allows you to make accurate structural assignments, even in the presence of high-intensity sample components present at the same retention time.
| Software Downloads | ||||
|---|---|---|---|---|
Name |
| Release Date | Release Notes | Download |
| 06.30.18 | Release Notes | Download | ||
| MetabolitePilot 2.0.4 Software License Terms | 05.10.18 | Download | ||
|
| All Software Downloads | ||