ProteinPilot software is a paradigm shift in protein identification and relative protein expression analysis for protein research. Combining sophisticated algorithms with an intuitive interface, you can now confidently identify more proteins and search large numbers of post translational modifications, without increasing search time or false positives. ProteinPilot software is compatible with all proteomics MS/MS systems via the.*mgf format. Get to know ProteinPilot software >

ProteinPilot software
The ProteinPilot software difference
Easy
The incredibly intuitive and easy to use interface is designed with biology in mind.
Powerful
Detect more peptides and PTMs than ever before with the Paragon algorithm.
Accurate
Identify the right list of proteins, not the longest list, with the Pro Group algorithm
In-depth protein identification made easy
Extensive peptide identification with every search
Search for hundreds of biological and other modifications, genetic variants and unexpected cleavages simultaneously with the Paragon algorithm, and avoid an explosion of false positives.
Industry-leading Pro Group
The embedded, industry-leading Pro Group algorithm for protein-grouping analysis enables you to quickly and easily achieve protein isoform and protein subset differentiation, as well as false positives suppression.
Easy on top, powerful underneath
The unique user-interface is designed with biology in mind. It allows sample information to be entered based on terms used by laboratory scientists, and the software intelligently determines the search parameters using the novel parameter translation layer function.
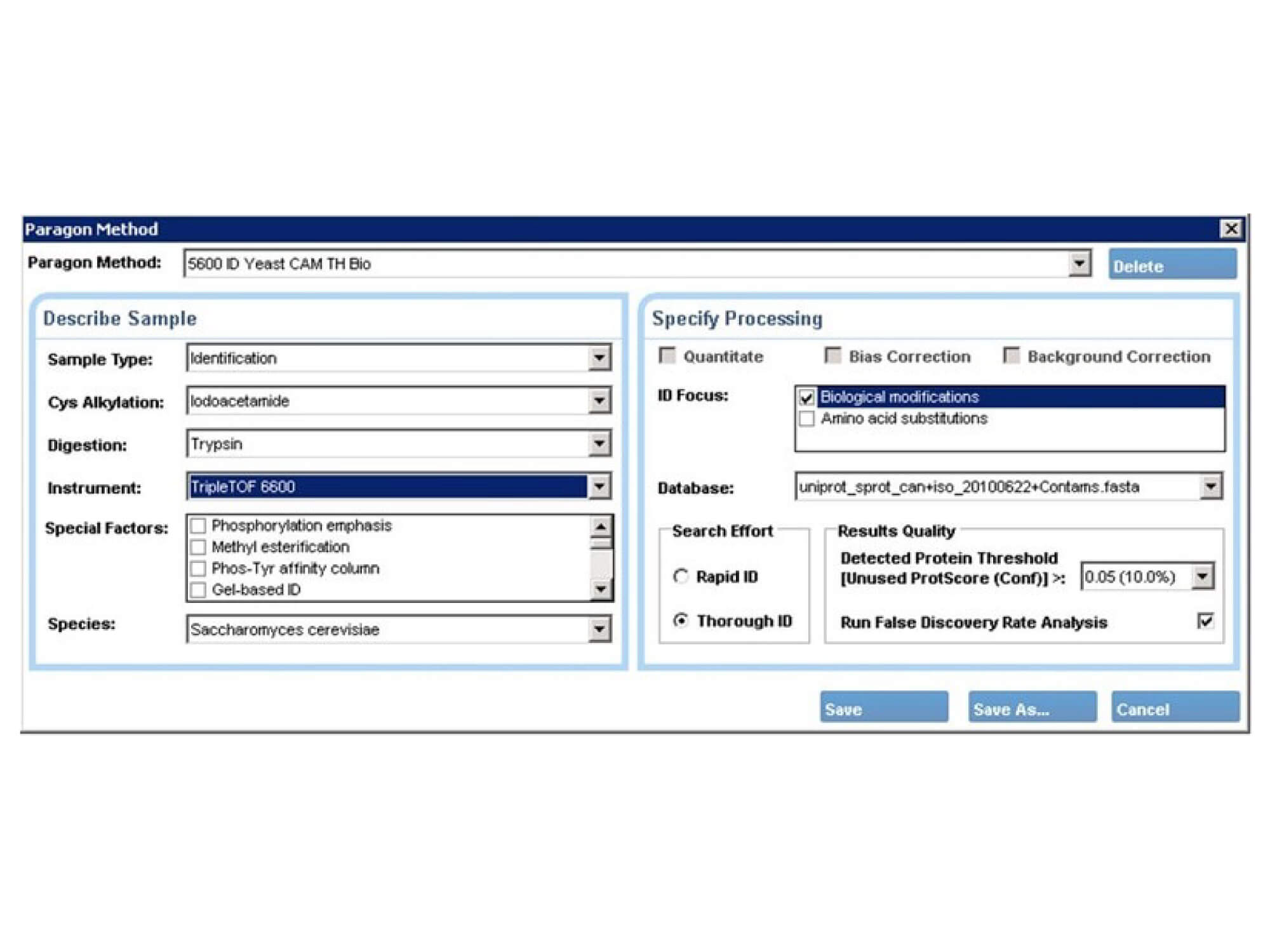
Top-quality identification results are delivered without the need for informatics expertise. When you have new exciting biology, the parameter translation layer can be edited by advanced users, to add new modifications or chemistries, to tune settings to optimize new workflows.
Valuable FDR analysis
Automatic and rigorous false discovery rate (FDR) analysis uses decoy database searching to provide high-quality, defensible protein identification results. A detailed report is generated with every search, with diagnostic dashboards to optimize sample prep and LC-MS acquisition conditions.
Optimized quantitative workflows
Benefit from optimized integrated support for iTRAQ reagents (4-plex and 8-plex) and numerous MS1 quantitation strategies (e.g. ICAT, SILAC, mTRAQ reagents) for protein expression analysis and biomarker discovery.
Software downloads
Name | File Size | Release Date | Release Notes | Download |
| 09.24.18 | Release Notes | Download | ||
| ProteinPilot Software v5.0.2 Software License Terms | 09.24.18 | Download | ||
| ProteinPilot 5.0.2 With No Sample Data and Fasta | 09.24.18 | Download | ||
| ProteinPilot Software 5.0.1 | 503 MB | 01.13.16 | Download | |
| ProteinPilot Software v5.0 Software License Terms | 02.24.14 | Download |
Resources
-
PDF
ProteinPilot 5.0.2 Software Release Notes
-
PDF
Template for Software HotFix Documentation
-
PDF
ProteinPilot Software Release Notes
-
PDF
ProteinPilot™ Software Release Notes
-
PDF
ProteinPilot software 5.0.2 EULA
-
PDF
Microsoft Word - EULA ProteinPilot v 5.0.DOC
-
PDF
Forensic Library 1720110-01
-
Technical note
Ultra-sensitive host cell protein detection using CESI-MS with SWATH acquisition
-
Technical note
iTRAQ to Investigate IκB Kinase Inhibition in Breast Cancer Cells
