Distinguishing oxidative impurities from ionizable lipids used in LNP formulations using electron activated dissociation
Achieving structural elucidation and distinction of oxidative isomer impurities of lipids with electron-activated dissociation (EAD) and the ZenoTOF 7600 system
Adam Crowe1, Nikita Jain1, Rehan Higgins1, Robert Proos2, Matthew Stone2 and Kerstin Pohl2
1Precision NanoSystems Inc., Canada; 2SCIEX, USA
Abstract
Here, the applicability of this novel fragmentation mode for the detailed characterization of lipids used for LNPs was tested using MC3 and its related impurities as a model. Within a single experiment, the exact locations of oxygen incorporation of 2 isomeric species and the double bond reduction of another related impurity were pinpointed using the unique fragment ions produced by EAD. This information can be used to determine drug efficacy and safety from formulated LNPs. Additionally, it can be used to aid rational design of new synthetic lipids.
Introduction
In this technical note, the comprehensive characterization of impurities from the ionizable lipid (6Z,9Z,28Z,31Z)-heptatriaconta-6,9,28,31-tetraene-19-yl-(dimethylamino)butanoate, commonly known as DLin-MC3-DMA (MC3), is presented. Deep structural elucidation, including the localization of different oxidation products and saturation of double bonds in MC3, was achieved using EAD.
Lipid nanoparticles (LNPs) that are comprised of ionizable lipids are used to deliver oligonucleotides to work as therapeutics or to stimulate the immune system, as in the initial mRNA-based COVID vaccines. A recent study reported that N-oxidation of ionizable lipids might lead to covalent modification of ribonucleotides and a loss of mRNA potency.1 To ensure product quality, detailed and sensitive characterization of the ionizable lipid and its related impurities is necessary. However, obtaining the level of detail needed is challenging with current liquid chromatography-mass spectrometry (LC-MS)-based methodologies. Collision-induced dissociation (CID) provides head group and acyl or alkyl chain sum composition information but does not provide structural details. Data from alternative fragmentation techniques can help localize double bond positions within acyl or alkyl chains but alternative methods suffer from inefficient fragmentation, especially for singly charged species. They require long duty cycles or collision cell modifications to allow for the introduction of ozone. Only EAD has efficiently provided complete characterization of different naturally occurring lipids in a single LC-MS run.2,3
Here, the applicability of this novel fragmentation mode for the detailed characterization of lipids used for LNPs was tested using MC3 and its related impurities as a model. Within a single experiment, the exact locations of oxygen incorporation of 2 isomeric species and the double bond reduction of another related impurity were pinpointed using the unique fragment ions produced by EAD. This information can be used to determine drug efficacy and safety from formulated LNPs. Additionally, it can be used to aid rational design of new synthetic lipids.
Key features of lipid impurity characterization with EAD
- Confident detection of low-level impurities with the ZenoTOF 7600 system and more than 5 orders of dynamic range
- Information-rich MS/MS spectra for definitive and distinct structural elucidation of related and isomeric singly charged lipid species with EAD
- Detection and identification of lipid impurities at levels below 0.01% relative abundance of the main peak by greatly enhanced fragment detection with the Zeno trap
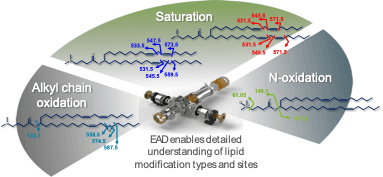
Figure 1. Location of bonds uniquely fragmented by EAD within the structure of MC3 and its related impurities. Arrows point to bonds in MC3 and its related impurities that were unique bond cleavages using EAD. These sites were used to localize specific sites of oxygen incorporation and double bonds to elucidate compound structures.
Methods
Sample preparation: A stock solution of MC3 (2 mg/mL) was diluted 1:10 in mobile phase A, consisting of 15:30:55, water/acetonitrile/methanol with 10mM ammonium acetate.
Chromatography: Two µL of the diluted MC3 sample (0.2 mg/mL) was injected onto an ExionLC AD system equipped with a reversed-phase column (C18, 1.7 µm, 2.1 × 150 mm). The column oven was set to 70°C. A total runtime of 27 min was used with a flow rate of 0.5 mL/min. Mobile phase A is described above and mobile phase B was 60:40, acetonitrile/methanol with 10mM ammonium acetate. Chromatographic conditions are described in Table 1.
Mass spectrometry: Data were acquired using SCIEX OS software on the ZenoTOF 7600 system in positive polarity. Data were collected from a single injection, using a combination of data-dependent acquisition (DDA) and a targeted approach that implemented an inclusion list. To compare the information generated from CID with EAD, both fragmentation techniques were used in separate experiments. Relevant MS parameters for the EAD method are described in Tables 2 and 3. CID fragmentation was acquired using a collision energy of 40 V and a 10 ms accumulation time.
Data processing: Structural elucidation was performed and relative quantification was determined using the Explorer and Analytics modules of SCIEX OS software, respectively.
Table 1. LC conditions.
Table 2. TOF MS and EAD MS/MS parameters.
Table 3. Inclusion list for MS/MS method.
Detection of MC3 and low abundance impurities
MC3 is the ionizable lipid used in LNP formulations for the therapeutic siRNA, patisiran. Like other ionizable lipids, its structure (Figure 1) contains a tertiary amine. The amine is on a 3-carbon head group that is bonded via an ester linkage to 2 separate but identical alkyl chains, each containing double bonds at C6 and C9. A preparation of MC3 was subjected to reversed-phase LC-MS analysis using the ZenoTOF 7600 system. Chromatographic separation showed a main peak (MP MC3) at 15.46 min (Figure 2, top). In addition, several low abundance impurity peaks were observed at less than 0.3% relative intensity of the MP at 13.1, 13.4 and ~1% at 16.9 min. Peaks at less than 0.01% relative intensity of the MP were also observed (data not shown), demonstrating the wide interscan dynamic range of the ZenoTOF 7600 system for lipid impurity analysis. Table 4 summarizes various impurities identified and their abundances, based on TOF MS peak area relative to the MP.
Figure 2. Structural confirmation of MC3 by LC-MS/MS using EAD. Top) Extracted ion chromatograms of the MC3 main peak and select detected impurities, shown in insets. Bottom left) TOF MS of the MC3 main peak and bottom right) MS/MS spectra of MC3 acquired by CID or EAD are shown. The insets in the MS/MS spectra demonstrate the richness of spectral depth that is generated from EAD but not CID.
Table 4. Compounds identified in the MC3 sample using EAD and their relative abundances based on TOF MS peak area.
Confirmation of MC3 structure
The m/z observed by TOF MS for the singly charged MP (Figure 2, bottom left) matched the theoretical m/z of MC3 within 1 ppm. This m/z was selected for fragmentation using either CID (Figure 2, middle right) or EAD (Figure 2, bottom right). Compared to CID, EAD provided a more information-rich spectrum, as evident by comparison of both insets. More than 100 peaks generated by EAD could be assigned to structures from in silico-derived fragments within less than 0.01 mDa mass error, compared to approximately 10 fragment ions generated from CID. The fragments generated by EAD were used to define discreet parts of MC3 to confirm its structure more confidently than with CID. For example, fragment ions at m/z 505.5, 531.5, 545.5 and 571.5 were used to confirm localization of double bonds at C6/C31 and C9/C28 (m/z were rounded to facilitate readability). Although CID and EAD both produced a product ion at m/z 132.1 that represents the dimethylamino butanoate head group, only EAD produced the complementary fragment ion at m/z 511.5 that represents the 2 identical alkyl chains. Overall, EAD could uniquely provide the fine structural details to confirm the exact structure of the MP as the singly charged MC3.
Site-specific localization of oxygen incorporation from 2 isomeric impurities
In addition to the MP, several impurity peaks were observed from the LC-MS chromatogram. Two peaks that eluted at 13.1 and 13.4 (Impurity 1 and Impurity 2, respectively) had the same precursor m/z of ~659 and isotopic distribution (Figure 3, top). An increase of ~16 amu was observed for these peaks compared to the MP MC3, indicating that these isomers each contained the addition of an oxygen atom. To elucidate the structures of these isomers, MS/MS was performed with either CID (Figure 3, bottom) or EAD (Figure 4). Despite the relatively low abundance of these impurity peaks (0.1-0.3% of the MP), high spectral quality was observed for both fragmentation modes, which can be attributed to the use of the Zeno trap.
The CID spectrum for Impurity 1 (Figure 3, left) provided the previously discussed m/z 132.1 of the head group, suggesting that the increase of ~16 amu occurred on the alkyl chains.
However, the position of the potential oxygen incorporation could not be determined based on the marginal MS/MS information derived from CID. Impurity 2 contained a unique peak at m/z 148.1 that was not observed in the MP MC3 MS/MS spectra. A peak at m/z 132.1 representing the dimethylamino butanoate head group was not observed. The increase of ~16 amu suggested that oxygen incorporation occurred somewhere within the head group but further information to confirm a specific position was missing in the CID spectrum.
In contrast to CID, EAD resulted in comprehensive fragmentation of both oxidated isomers (Figure 4). These results were similar to those observed for the MP MC3, further demonstrating the spectral quality of EAD for low abundance species and the excellent sensitivity of the ZenoTOF 7600 system.
The fragment ion at m/z 132.1 of Impurity 1 in the EAD spectrum suggested that oxygen incorporation resided on the alkyl chains and not within the dimethylamino butanoate head group. In addition, several unique fragment ions between m/z 500-600 (Figure 4, top) were generated with EAD, that were not found for the MP MC3. The CID spectra of Impurity 1 did not contain these ions or other ions to define the structural details of the alkyl chains (Figure 3, bottom). The presence of these ions could only be explained by the addition of oxygen between C5/C32 and C6/C31. From these data, an epoxide at this position in the structure of Impurity 1 was proposed (Figure 4, top). The lack of diagnostic ions at m/z 61.05 and m/z 511.5, described below, further confirmed that the addition of oxygen did not reside within the head group.
Both CID and EAD product ion spectra for Impurity 2 contained a unique peak at m/z 148.1 that was not observed in the MP MC3 product ion spectra. A peak at m/z 132.1 representing the dimethylamino butanoate head group was not observed. The increase of ~16 amu suggested an oxygen incorporation within the structure of the dimethylamino butanoate head group. The EAD spectra of Impurity 2 showed the presence of the m/z 511.5 ion, which was also observed in the MP MC3 EAD spectra (data not shown). The lack of any mass shift by ~16 amu for this ion and other ions assigned to other structures within the alkyl chains further substantiated oxygen incorporation within the head group. A unique ion at m/z 61.05 within the EAD fragment ion spectra of Impurity 2 was also observed. This ion could only be explained by a structure in which oxygen was incorporated within the dimethylamino group itself, thereby greatly restricting possible structures that could represent Impurity 2. As such, Impurity 2 was identified as the N-oxide species. Only EAD could provide this level of detail for both low level impurities.
Figure 3. Data from 2 oxidative impurity isomers of MC3. Top) TOF MS data of 2 MC3-related impurities that elute before MP MC3 at 13.1 and 13.4 min (see Figure 2). Both impurities show highly similar MS1 data with an increase in ~16 amu compared to MP MC3. The earlier elution time and the ~16 amu mass shift suggest oxygen incorporation. Bottom) CID MS/MS spectra of the 2 impurities. The insets show the limited fragment ion data acquired by CID. For both impurities, the exact location of the suggested oxygen incorporation remained unclear using CID MS/MS. For readability purposes, fragment m/z values were rounded.
Figure 4. Distinction of 2 oxidative impurity isomers of MC3 by EAD. The EAD MS/MS spectra and the unique ions that were generated and used to identify and discriminate between the 2 oxygenated isomers are shown. Specific ions observed at m/z 558.5, 574.5 and 587.5 show oxygen incorporation at C6/C31 of the alkyl chain for Impurity 1 (blue circles, top). The specific fragment ion at m/z 61.05 showed that oxygen incorporation was in the tertiary amine (N-oxide) of Impurity 2 (green circles, bottom). For readability purposes, fragment m/z values were rounded.
Site-specific determination of a saturation impurity of MC3 using EAD
In addition to oxygen incorporation, other impurities were observed within the chromatogram of the MC3 preparation. The impurity peak that eluted at 16.9 min showed a gain of ~2 amu compared to the MP MC3. Fragmentation of this impurity peak by EAD resulted in unique ions at m/z 533.5, 547.5, 559.5 and 573.5 (encircled m/z in Figure 5). These ions had an increase of ~2 amu compared to ions from the MP MC3 data. The EAD data for MC3 showed a unique ion at m/z 571.5 (encircled m/z Figure 5). Common fragment ions at m/z 531.5 and 545.5 were observed for both species. Taken together, these data demonstrated that the impurity was a result of 1 double bond reduction by the gain of 2 protons specifically at the C6/C31 position (Figure 5).
Figure 5. Specific localization of a reduced double bond on an MC3 impurity using EAD. Overlay of EAD MS/MS spectra of the main peak MC3 (red) and impurity peak that eluted at 16.9 min (blue; see Figure 2). Unique ion pairs at m/z 558.5/559.5 and 572.5/573.5 indicated the location of 1 double bond reduction at C6/C31. For readability purposes, fragment m/z values were rounded. Unique fragment ions for each species are encircled.
Outlook
EAD was used to characterize the structures of the ionizable lipid MC3 and low-level related impurities. This was achieved in a single-injection LC-MS/MS experiment that used a 20 min gradient. Compared to other fragmentation techniques, the resulting product ion data were comprehensive and provided in-depth structural detail throughout the lipid species analyzed. Data from EAD were used to distinguish structural isomers of 2 MC3 oxidative impurities and determine specific sites of oxygen incorporation. These data were also used to localize specific sites of double bonds and related saturation impurities.
Complete structural analysis of lipids using EAD is not limited to MC3. It has also been used to provide complete structural details of glycerophospholipids2,3 and other ionizable lipids and their related impurities. This technique can be used to provide rapid, sensitive and detailed structural analysis of lipids within LNP formulations for liability studies and for raw material testing. No other MS-based fragmentation technique can provide this level of structural detail for singly charged species. Overall, the information provided by EAD can enhance product quality and safety of LNP-formulated products. Additionally, it can help guide synthesis and development of novel ionizable lipids that have higher quality, stability and therapeutic efficacy to further unlock the potential of RNA-based therapeutics and vaccines.
Conclusions
- Increased efficiency by obtaining in-depth structural characterization of singly charged, ionizable lipids and related impurities within a single LC-MS/MS run
- Improved risk assessment of raw material and formulated LNPs through structural distinction and site-specific localization of molecular oxygen incorporation of MC3 structural isomer impurities with Zeno EAD
- Increased confidence in product quality assessment by determination of exact double bond locations in MC3 and specific sites of double bond saturation from related impurities using Zeno EAD
- Decreased risk of missing critical low abundance impurities due to great interscan dynamic range and relative quantification capabilities of the ZenoTOF 7600 system
- Accelerating lipid formulation assessment by extension of the Zeno EAD method to other lipid species for detailed structural elucidation of main compound and impurities
References
1. Packer, M., Gyawali, D., Yerabolu, R. et al. 2021: A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat Commun 12, 6777.
2. Complete structural elucidation of lipids in a single experiment using electron activated dissociation (EAD). SCIEX technical note, RUO-MKT-02-13050-B.
3. Systematic determination of lipid structure using electron activated dissociation (EAD). SCIEX technical note, RUO-MKT-02-14182-A.
 Click to enlarge
Click to enlarge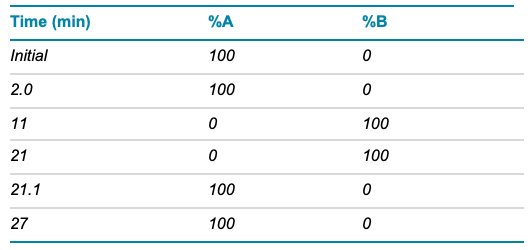 Click to enlarge
Click to enlarge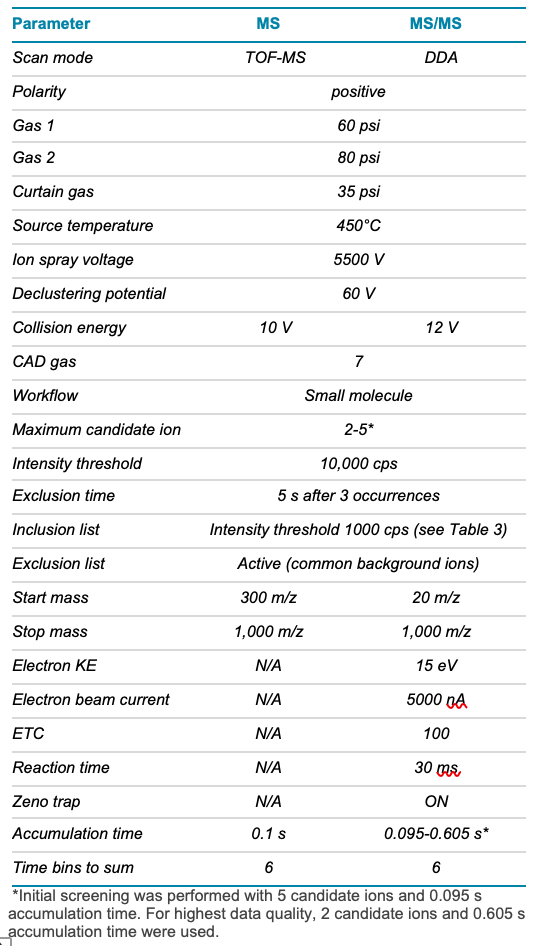 Click to enlarge
Click to enlarge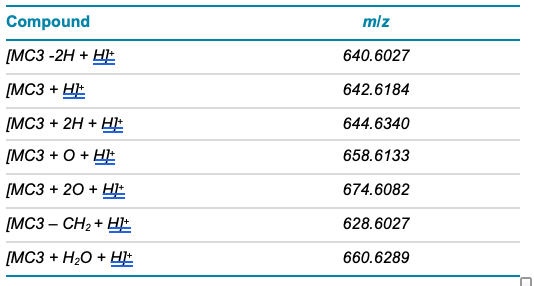 Click to enlarge
Click to enlarge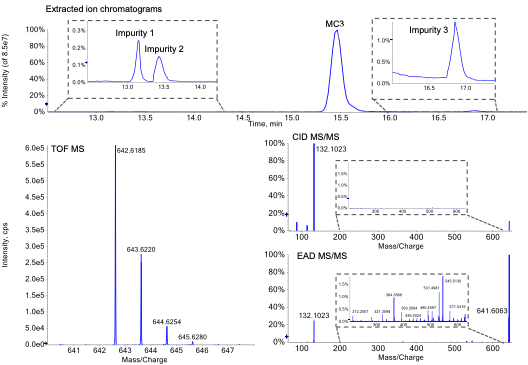 Click to enlarge
Click to enlarge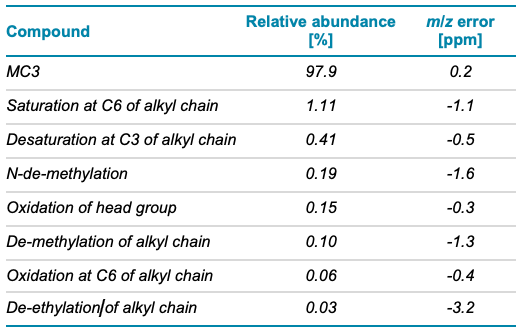 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge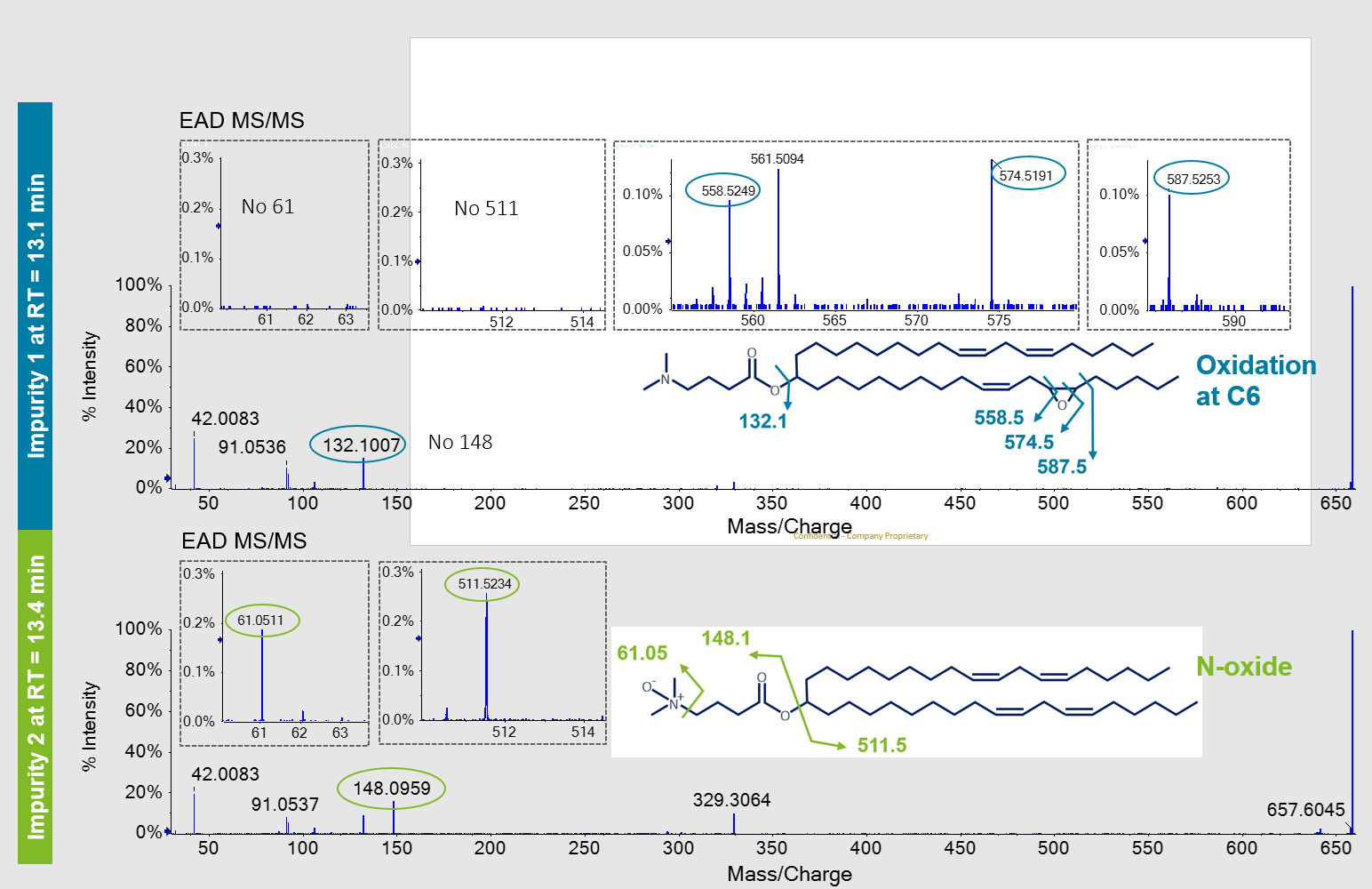 Click to enlarge
Click to enlarge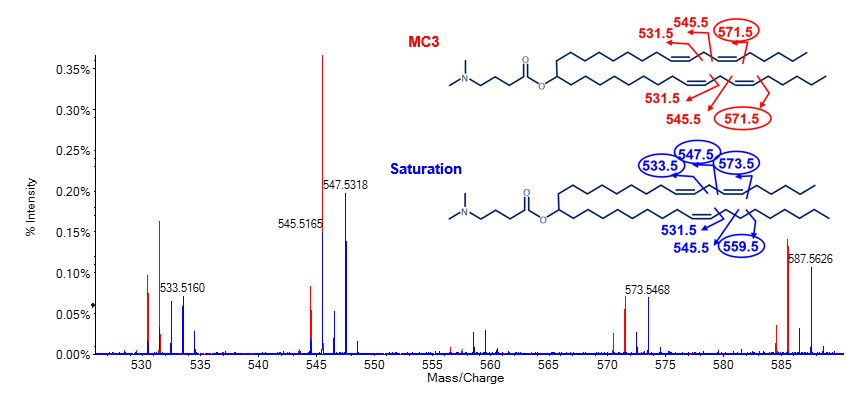 Click to enlarge
Click to enlarge