Abstract
In this technical note, the SCIEX X500 QTOF system was used to analyze PFAS in human serum. An unbiased and non-targeted workflow with automatic library matching allowed for wider, comprehensive sample analysis. Additionally, a streamlined sample preparation strategy maintained sample integrity while providing greater laboratory efficiency.

Introduction
Per- and poly-fluorinated alkyl substances (PFAS) are used extensively in consumer and industrial products.1 Although they have been highly beneficial to society, it is now widely recognized that health effects such as immunosuppression and serum lipid alteration are linked to continued exposure and accumulation of PFAS in the body.2
Evidence for organic fluorine compounds in human serum was first discovered over 50 years ago,3 however the LC-MS/MS techniques necessary to identify them were not widely available until the early 2000s. Targeted LC-MS/MS strategies for PFAS detection provide high confidence and accuracy, but only detect a limited subset of compounds. In contrast, non-targeted strategies using high resolution accurate mass instruments provide greater coverage and can be used to identify unknown compounds. For example, MS/MS spectra can be matched to library databases for identification.
In this technical note, the SCIEX X500 series QTOF system was used to analyze PFAS in human serum and plasma. An unbiased and non-targeted workflow with automatic library matching allowed for wide analyte coverage. Additionally, a streamlined sample preparation strategy provided greater laboratory efficiency, while maintaining sample integrity.
Key features of SWATH acquisition for PFAS identification
- High resolution accurate mass SWATH acquisition data provide multiple levels of evidence for identification and compound confirmation (i.e. mass error, isotope pattern, MS/MS library matching)
- High resolution PFAS MS/MS library covers negatively charged, positively charged and zwitterionic compound classes and allows users to add custom compounds
- SCIEX OS allows the user to add MS/MS spectra from authentic standards or sample data for those compounds not yet in the library such as novel PFAS compounds
Methods
Sample preparation: Five pooled samples were prepared from discarded serum and/or plasma specimens. Samples (0.5 mL) were denatured using 2 mL of acetonitrile with 1% formic acid and then centrifuged. The supernatant was passed through a Captiva EMR-lipid cartridge (300 mg), the eluent evaporated under a nitrogen stream and the sample reconstituted in 200 µL of 1:1 water: methanol (1% formic acid).
Analytical standards: The EPA 533 PFAS analyte list was used to build the initial suspect screening list. Analytical standards were purchased from Wellington Laboratories (Guelph, ON, Canada) and analyzed to confirm retention times.
Chromatography: Chromatography was performed using an ExionLC system. Analytes were separated using a Phenomenex Gemini C18 column (110 Å, 100 x 3 mm, 3 μm particle size) using gradient conditions and a flow rate of 0.5 mL/min. A delay column was used to separate PFAS compounds originating from the LC. The mobile phases were water (A) and methanol (B), both with 10 mM ammonium acetate. The column oven was 40°C and the injection volume was 10 µL. Initial conditions were 20% B, held for 1 min and then increased to 99% B over 3.5 min.
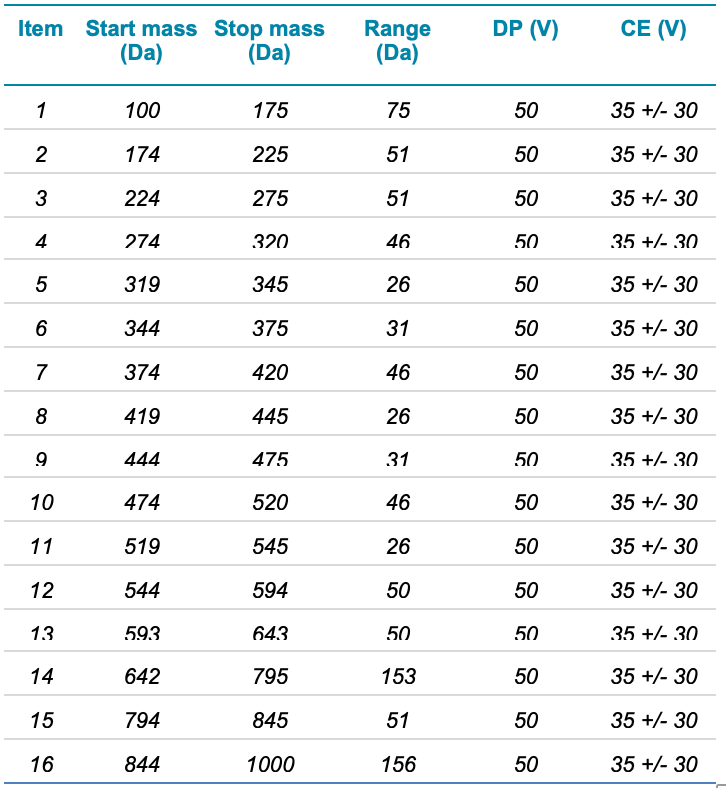
Mass spectrometry: Analysis was performed on the X500 series QTOF system with the Turbo V ion source using electrospray ionization (ESI) in negative ion mode. Data were collected using SWATH acquisition mode with a TOF MS experiment followed by 16 TOF MS/MS experiments using Q1 windows spanning 100 to 1000 Da. The TOF MS and each MS/MS experiment used an accumulation time of 50 ms, for a total cycle time of 0.85 s. Table 1 lists the variable Q1 windows used for SWATH acquisition and other related parameters. SWATH acquisition windows were carefully chosen to avoid overlap with co-eluting analytes, for example perfluorononanoic acid (PFNA) and perfluorooctane sulfonate (PFOS). The TOF MS scan ranged from 100 to 1000 Da using DP=-50 V and CE=-5 V. The source and gas conditions were: GS1= 60, GS2= 60, CUR= 35, CAD= 10, TEM= 500°C, ISV = -4500 V.
Data processing: Data acquisition and data processing were performed using Analytics within the SCIEX OS software. Analytics links the precursor extracted ion chromatogram (XIC) with the appropriate MS/MS fragmentation pattern. Library matching was automatically performed using the SCIEX Fluorochemical HR-MS/MS library 2.0, which contains 252 PFAS compounds. These compounds cover negative, positive and zwitterionic compound classes, and include both legacy and novel PFAS compounds, such as those originating from AFFF (aqueous film forming foam) and AFFF-impacted water.
Reducing bias through study design
There are approximately 5000 PFAS compounds used in global commerce. For this reason, most people in the world have been exposed to and have measurable concentrations of PFAS compounds in their blood.2,4 It has been shown that approximately 95% of the population have PFAS in their blood, and in particular PFOS (perfluorooctane sulfonic acid) and PFOA (perfluorooctanoic acid).
Serum is a complex matrix that contains many compounds that can interfere with PFAS detection and identification.5 As a result, serum samples often undergo preparation specifically intended to reduce these interferences. These preparation strategies, however, can bias both the overall composition of the final extract, as well as the concentrations of individual PFAS compounds. For this study, a simplified sample preparation, consisting of only denaturation, centrifugation and highly selective lipid/matrix removal, was used to reduce bias induced by sample clean-up and handling.
It is now recognized that there are many PFAS “unknowns” that are not detected using traditional targeted analyses. In fact, the proportion of undetected PFAS unknowns is increasing in recent years potentially due to the introduction of replacement compounds.6 Thus, non-targeted strategies using high resolution accurate mass instruments are typically now used for the confidence they confer in the identification of PFAS compounds. Because of the high number of PFAS compounds and overlap with matrix interferences (which are potentially even more profound with the simplified sample preparation used here), the addition of high resolution accurate mass analysis is necessary to distinguish between and correctly identify the many PFAS and confounding matrix compounds present in samples.5
SWATH acquisition is a type of data independent acquisition (DIA), that uses an unbiased approach for the acquisition of MS/MS data from complex samples. In contrast to the more traditional information dependent acquisition (IDA), which relies on a set of preconfigured criteria for precursor selection and subsequent MS/MS acquisition, SWATH acquisition captures MS/MS spectra on all detectable compounds. SWATH acquisition uses a wide Q1 isolation window to step across the mass range to sequentially acquire MS/MS data for every precursor ion. Once the data have been acquired, they are stored as a digital archive of the sample which can also be used for retrospective analysis, if necessary. Data processing algorithms then deconvolute the fragment data to assign precursor ions to product ions.
In this study, 16 variable width SWATH acquisition windows were used to step across the entire precursor mass range to fragment all PFAS precursors (Table 1). Variable width windows were used to reduce the total number of precursors in certain densely populated regions and thereby reduce the complexity of the MS/MS spectra and increase specificity. Once the data were collected, the entire data file was subjected to suspect screening to determine whether specific compounds were present in the samples. In addition, MS/MS library matching was performed for compound confirmation.
Results
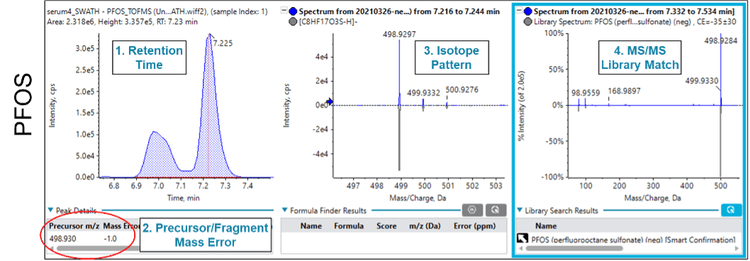
Figure 1 shows the SWATH acquisition data for perfluorooctane sulfonate (PFOS) identified in the serum sample. Multiple lines of evidence support the identification of PFOS including:
- the retention time ✔︎
- the accurate precursor mass with low mass error ✔︎
- the isotope pattern observed for the precursor mass ✔︎
- the library match to the MS/MS fragment data ✔︎
Figures 2, 3 and 4 show similar data for perfluorobutanoic acid (PFBA), PFNA and perfluoroheptane sulfonic acid (PFHpS), which were also identified in the serum sample. The multiple lines of evidence outlined above confirm the presence of these compounds in the serum sample.
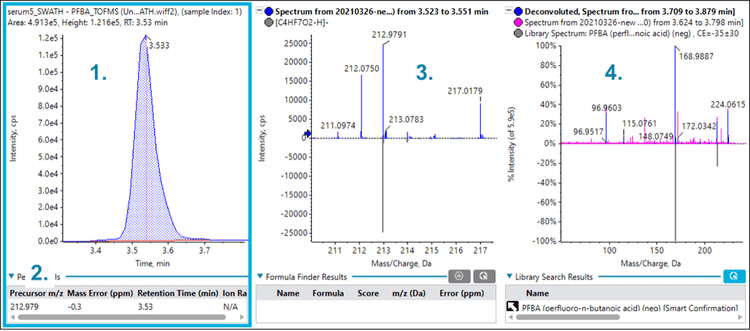
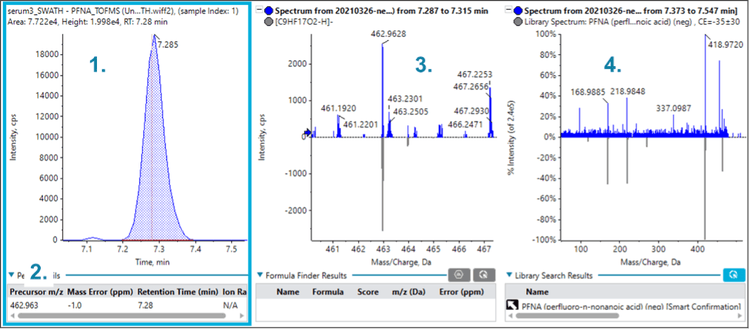
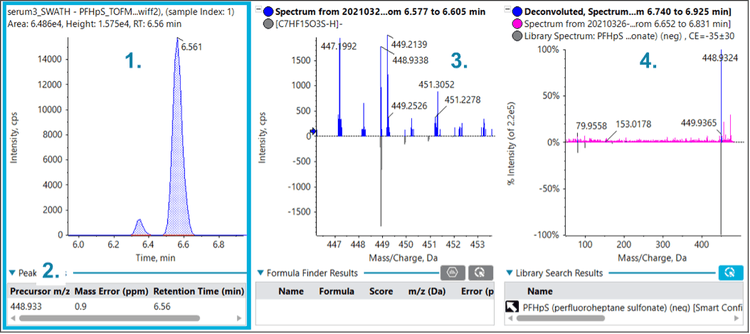
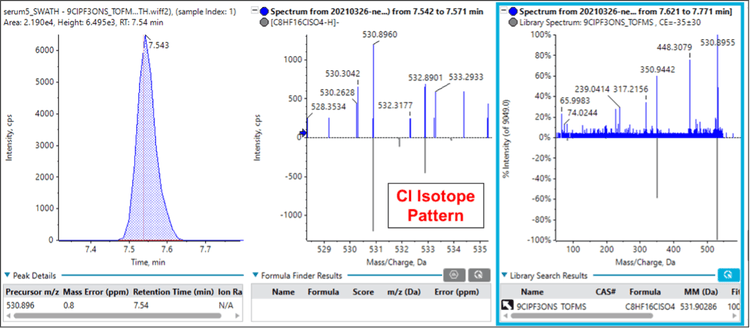
Figure 5 shows an interesting situation in which a compound was not initially identified – despite being included on the suspect screening list – since the fluorochemical library did not have a MS/MS spectrum for this chemical. Careful examination of the TOF MS isotope pattern indicated that the compound likely contained a chlorine atom. Note that because of the wider Q1 isolation windows, the full isotope pattern was preserved in the MS/MS data, enabling the detection of the distinctive isotope pattern for Cl. Further, the precursor mass matched that of the PFOS replacement compound, 9-chlorohexadecafluoro-3-oxanonane-1-sulfonic acid (9ClPF3ONS) with a low mass error.
An authentic standard of 9ClPF3ONS was obtained and run using SWATH acquisition to confirm the identity of the unknown PFAS compound. The MS/MS spectrum generated for 9ClPF3ONS was added to the fluorochemical library. Re-processing the serum sample data and comparing it against the updated fluorochemical library now showed a positive identification of the unknown as 9ClPF3ONS. Notably, this positive identification of 9ClPF3ONS was accomplished using previously collected data, as SWATH acquisition creates a digital archive of the sample. This retrospective analysis could prove valuable in the future as more novel PFAS compounds are reported.
Conclusions
With the high number of known PFAS compounds in existence today and the growing number of new and unknown PFAS, a non-targeted LC-MS/MS method provides the most comprehensive strategy for PFAS identification. SWATH acquisition is a non-targeted data independent acquisition (DIA) workflow and therefore provides an unbiased approach for capturing MS/MS data for all detectable PFAS when utilizing the appropriate polarity and ionization technique. SWATH acquisition provides high resolution accurate mass data on precursors and product ions, isotope information, retention time and MS/MS library matching to help identify and confirm compounds from highly complex matrices. Moreover, the crude serum sample preparation used here minimizes losses from extensive sample handling, preserving the integrity of the sample and allowing data capture on as many known and unknown PFAS compounds as possible.
The ability of SCIEX OS software to allow the user to rapidly add MS/MS fragmentation spectra into new or existing libraries results in enhanced flexibility for compound identification during non-target analysis. Coupled with the in-depth retrospective data processing possible with SWATH acquisition, this workflow will allow users to easily adapt as new PFAS compounds are discovered.
References
- Buck RC, et. al., (2011) Perfluoroalkyl and Polyfluoroalkyl Substances in the Environment: Terminology, Classification, and Origins. Integr. Environ. Assess. Manag. 7(4): 513-541.
- Sunderland EM, et. al., (2019) A Review of the Pathways of Human Exposure to Poly- and Perfluoroalkyl Substances (PFASs) and Present Understanding of Health Effects. J. Expo. Sci. Environ. Epidemiol. 29(2): 131-147.
- Taves DR, (1968) Evidence that there are Two Forms of Fluoride in Human Serum. Nature 217: 1050–1051.
- De Silva AO, et. al., (2021) PFAS Exposure Pathways for Humans and Wildlife: A Synthesis of Current Knowledge and Key Gaps in Understanding. Environ. Toxicol. Chem. 2021(40): 631-657.
- Reducing PFAS interferences in human plasma and serum with accurate mass spectrometry. SCIEX technical note, RUO-MKT-02-13393-A.
- Miaz LT, et. al., (2020) Temporal trends of suspect- and target-per/polyfluoroalkyl substances (PFAS), extractable organic fluorine (EOF) and total fluorine (TF) in pooled serum from first-time mothers in Uppsala, Sweden, 1996–2017. Environ. Sci.: Processes Impacts 22: 1071-1083.