- Home
- Life Science Research
- Metabolomics
- Extending metabolome coverage - Untargeted metabolomic workflows - NIST library
Extending metabolome coverage
Using the licensed NIST ‘17 MS/MS spectral library for untargeted metabolomics workflows
Oscar G. Cabrices and Baljit K. Ubhi
SCIEX, USA
Overview
Confident metabolite identification is a major bottleneck in the field of untargeted metabolomics today. To help alleviate this there is a need for larger libraries and databases of spectral compounds which can aid the metabolite identification process. The SCIEX All-In-One High Resolution MS/MS Spectral Library enables accurate compound detection and identification through library spectral matching. In combination with the licensed NIST ’17 MS/MS Library, the provides spectra for over 17000 compounds including human and plant metabolites, sugars, glycans and natural products commonly investigated in metabolomics research for complex samples, including blood, urine and tissues.

This library is for use with for use with the X500R QTOF System powered by SCIEX OS Software and also compatible for use with SCIEX TripleTOF® and QTRAP® Systems with SCIEX OS Software and LibraryView™ Software.
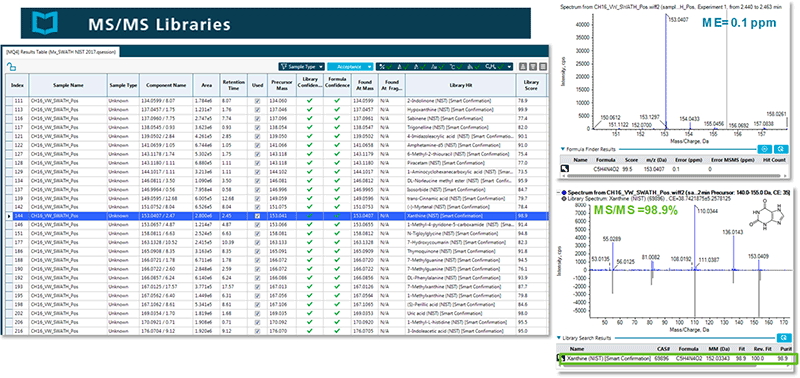
Figure 1 shows an example of an extracted human urine sample from a pre-classified prostate cancer study. In this example the metabolite Xanthine is highlighted. The accurate identification (MS/MS Library Score = 98.9%, Mass Error = 0.1 ppm) of this analyte was possible through the addition of the licensed NIST ’17 MS/MS Spectral Library to the SCIEX All-In-One High Resolution MS/MS Spectral Library.
Figure 1. Gain extended metabolome coverage with the addition of the licensed NIST ’17 high resolution MS/MS spectral library. Identify any metabolites found in the samples by reviewing the confidence scores for the MS1 mass error, retention time, isotope fidelity and the MS/MS. Xanthine (highlighted in the results table) is a purine base found in most body tissues and fluids, certain plants, and some urinary calculi. It is an intermediate in the degradation of adenosine monophosphate to uric acid, being formed by oxidation of hypoxanthine. This compound is associated with Lesch-Nyhan syndrome which leads to kidney problems through buildup of uric acid (Source: www.hmdb.ca).
References
- Improved metabolite identification using data independent analysis for metabolomics. SCIEX techincal note RUO-MKT-02-10617-A.
 Click to enlarge
Click to enlarge