Abstract
This technical note highlights an electron activated dissociation (EAD)-based middle-down workflow with controlled sample preparation to achieve high sequence coverage (88%) of the light chain (LC) and confident mapping of intra-chain disulfide linkages in a single injection.
Introduction
Middle-down mass spectrometry (MS) combines the benefits of bottom-up and top-down MS.1-8 It provides high sequence coverage of biotherapeutics with simple sample preparation, a low degree of artificial modification, no disulfide bond scrambling and easy data interpretation. Traditional middle-down MS workflows often involve multiple fragmentation techniques and/or injections with extensive method optimization to achieve high sequence coverage.7,8 One of the challenges with middle-down MS is the lack of fragmentation in the middle of a subunit sequence. This limitation can be overcome by applying EAD to disulfide-linked subunits.5,6 The application of EAD leads to complementary fragmentation patterns of the fully reduced and disulfide-linked subunits.5,6 The combined EAD results of the fully reduced and disulfide-linked subunits provided high sequence coverage (85- 93%) of mAb subunits in 2 injections.6 In this work, an EAD-based middle-down workflow with controlled sample preparation is presented to extend the capability of this powerful workflow for biotherapeutic characterization. The controlled denaturation and reduction of NISTmAb leads to the co-presence of the fully reduced and disulfide-linked LC subunits in the same sample. Targeted EAD fragmentation of these subunits provides high sequence coverage and high-confidence disulfide bond mapping from a single injection (Figure 1).
Key features of the EAD-based middle-down workflow for biotherapeutic characterization
- High sequence coverage in a single injection: 88% sequence coverage of the LC subunit in a single injection
- High-confidence disulfide bond mapping: EAD results in a distinctive fragmentation pattern of disulfide-linked subunits for rapid intra-chain disulfide bond mapping with minimal false positives
- Accurate localization of labile PTMs: Labile PTMs are preserved in the EAD fragments
- Streamlined and easy to implement: The workflow requires minimal method optimization and is streamlined from data acquisition to results review
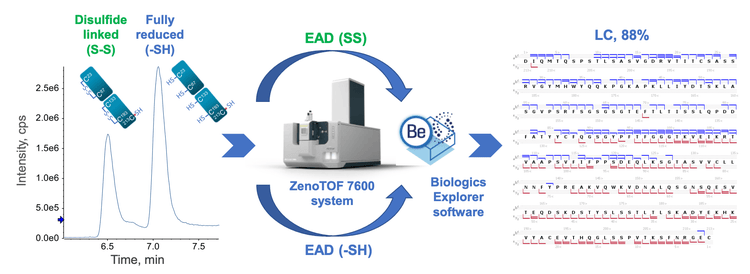
Methods
Samples preparation: An aliquot of 10 µg/µL NISTmAb was mixed with guanidine hydrochloride (Gnd) and dithiothreitol (DTT) at a final concentration of 2M and 50mM, respectively. The mixture was incubated at 45°C for 30 minutes to generate LC and HC subunits. 4 µL of the final solution (~4 µg) was injected for LC-MS analysis immediately following the completion of the incubation.
Chromatography: The subunits of NISTmAb were separated using an ACQUITY UPLC Protein BEH C4 column (2.1 × 50 mm, 1.7 µm, 300 Å, Waters). The LC gradients used for the subunit separation are shown in Table 1. A flow rate of 0.3 mL/min was used for all LC runs. The column was kept at 60°C in the column oven of an ExionLC AD system (SCIEX). Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile.
Mass spectrometry: MRMHR EAD experiments were performed in SCIEX OS software using the ZenoTOF 7600 system (SCIEX). 2 charge states of the fully reduced (24+ and 19+) and disulfide-linked (18+ and 15+) LC subunits were targeted for EAD fragmentation in this work. The key TOF MS and MRMHR settings used are listed in Table 2 and 3, respectively.
Data processing: MRMHR data were analyzed using a middle-down workflow template in Biologics Explorer software. EAD spectra of the disulfide-linked LC subunit were processed separately for disulfide bond mapping, or jointly with those of the fully reduced counterpart in the same data file to obtain high sequence coverage.
EAD-based middle-down analysis of the fully reduced and disulfide-linked subunits
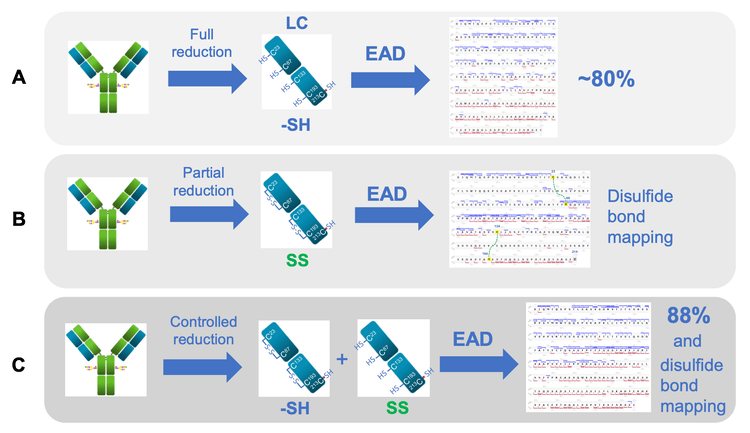
It was demonstrated previously that the EAD-based middle-down workflow is a highly effective approach for obtaining high sequence coverage and confident disulfide-bond mapping of antibody subunits (Figure 2).1-6 EAD leads to complementary fragmentation patterns from the fully reduced and disulfide-linked subunits.5,6 EAD fragmentation of the fully reduced subunits occurs preferentially in the regions near the 2 termini (Figure 2A).1-4 By comparison, EAD of the disulfide-linked subunits results in a characteristic fragmentation pattern, where the middle of the sequence outside the disulfide-forming areas is extensively cleaved (Figure 2B). 5,6 The distinctive fragmentation pattern of the disulfide-linked subunits provides additional sequence coverage and confident mapping of intra-chain disulfide linkages without false positives.5,6 By combining EAD results of the fully reduced and disulfide-linked subunits high sequence coverages (85-93%) of antibody subunits can be obtained in 2 injections.6
To further streamline the EAD-based workflow, the condition of sample preparation was fine-tuned to generate the fully reduced and disulfide-linked LC subunits in the same sample. EAD fragmentation of these 2 forms of the LC subunit provided high sequence coverage (88%) and rapid disulfide mapping in a single injection (Figure 2C).
Co-formation of the fully reduced and disulfidelinked subunits
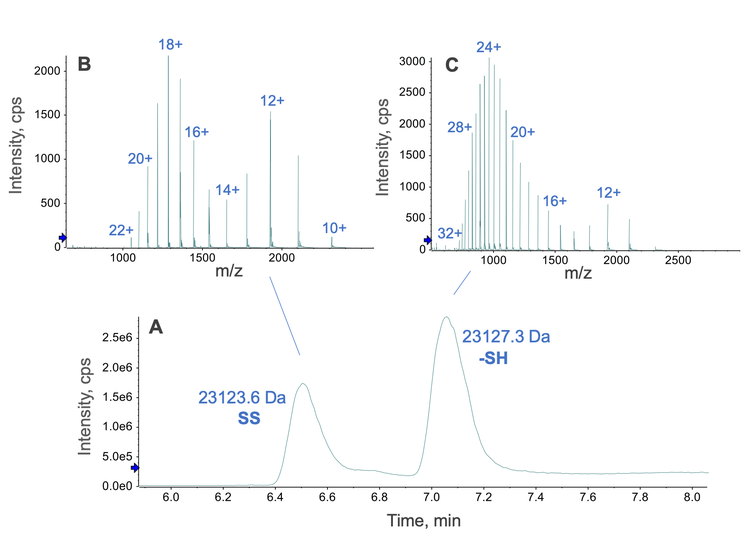
Fine-tuning the denaturation and reduction conditions produced the fully reduced and disulfide-linked LC subunits of NISTmAb in the same sample. The concentration of Gnd, the temperature and duration of incubation were the key factors in determining the ratio of the fully reduced vs. disulfide-linked subunits. In this study, a final concentration of 2M Gnd and an incubation temperature and time of 45°C and 30 minutes, respectively, led to the co-formation of the fully reduced and disulfide-linked LC subunits of NISTmAb (Figure 3). These 2 forms of the LC subunit can be chromatographically resolved (Figure 3A). The mass spectra of the 2 species (Figure 3B and 3C) show a distinct difference in charge state distribution. The disulfide-linked LC subunit carried fewer charges on average compared to the fully reduced counterpart due to a lesser degree of sequence unfolding because of the presence of 2 intra-chain disulfide bonds. Automated protein deconvolution performed using Biologics Explorer software confirmed the masses of the fully reduced (23,127.3 Da) and disulfide-linked (23,123.6 Da) LC subunits. 2 charge states from each form of the LC subunit were targeted for EAD fragmentation in a single MRMHR experiment. The processing of these EAD data separately or jointly led to high sequence coverage and rapid confirmation of intra-chain disulfide linkages for the LC subunit, as described below.
Achieving high sequence coverage and confident disulfide bond mapping of the LC subunit from a single injection
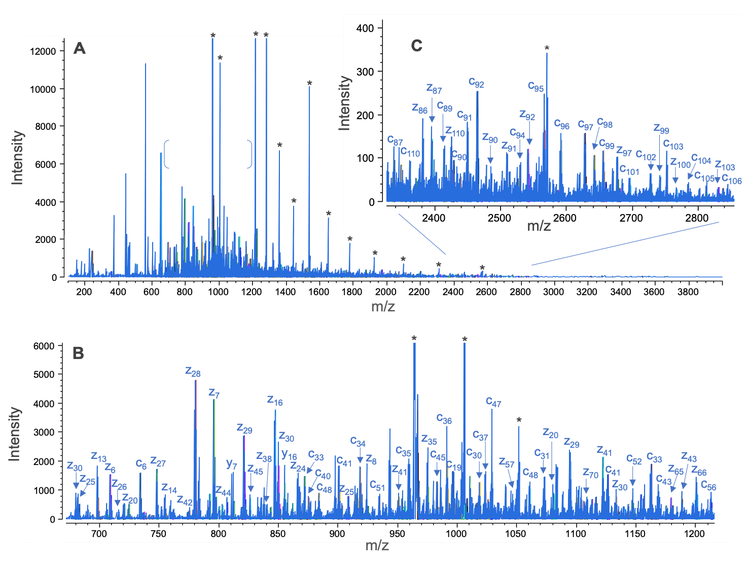
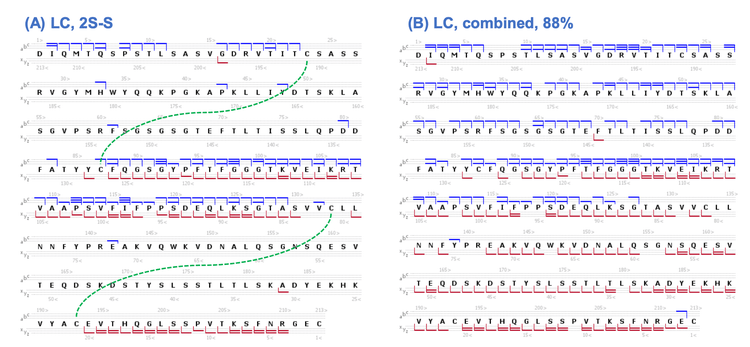
The formation of the fully reduced and disulfide-linked LC subunits of NISTmAb in the same sample enabled the EAD fragmentation of these 2 species in a single injection. The combined EAD spectrum of the 2 LC forms is composed of rich fragments across a wide m/z range of 100-4,000 (Figure 4A). Highlighted in Figures 4B and 4C are the zoom-ins of 2 selected spectral regions in the low and high m/z range where a large number of c/z fragments were detected. An abundant level of c/z fragments was also present in the rest of the spectral regions (data not shown). EAD spectra of the fully reduced and disulfide-linked LC subunits can be processed separately or jointly―depending on the RT range restriction―using an intuitive middle-down workflow template within Biologics Explorer software. The interpretation of the EAD result of the disulfide-linked LC subunit led to a characteristic sequence coverage that is indicative of 2 intra-chain disulfide linkages (Figure 5A), in agreement with the previous results.5,6 The analysis of the combined EAD data from the 2 species produced high sequence coverage (88%) of the LC subunit in a single injection (Figure 5B).
In summary, an EAD-based middle-down workflow with controlled sample preparation was developed to provide high sequence coverage and disulfide bond mapping of antibody subunits in a single injection. The controlled sample preparation led to the formation of the fully reduced and disulfide-linked LC subunits in the same sample. The complementary EAD fragmentation patterns of these 2 species resulted in high sequence coverage (88%) of the LC subunit. In addition, EAD enabled the confident determination of 2 intra-chain disulfide linkages in the disulfide-linked subunit. The strategy described in this technical note can improve the efficiency and effectiveness of using the middle-down workflow for the comprehensive characterization of biotherapeutics with increasing diversity and complexity.
Conclusion
- An EAD-based middle-down workflow with controlled sample preparation provides high sequence coverage (88%) of the LC subunit of NISTmAb in a single injection
- The characteristic fragmentation pattern offered by EAD for the disulfide-linked subunits enables confident determination of intra-chain disulfide bonds
- Biologics Explorer software offers easy-to-use middle-down workflow templates and powerful visualization tools for improved user experience with data analysis
References
- A streamlined single-injection middle-down workflow using electron activated dissociation (EAD) for biotherapeutics characterization. SCIEX technical note, MKT-26997-A.
- Obtaining high sequence coverage and confident posttranslational modification (PTM) analysis of biotherapeutics using an electron activated dissociation (EAD)-based middledown workflow. SCIEX technical note, MKT-27223-A.
- Comparative analysis of biotherapeutics using an electronactivated dissociation (EAD)-based middle-down workflow. SCIEX technical note, MKT-27427-A.
- Confident sequence analysis of a trispecific antibody using an electron-activated dissociation (EAD)-based middle-down workflow. SCIEX technical note, MKT-27784-A.
- High-confidence disulfide bond mapping of biotherapeutics using an electron-activated dissociation (EAD)-based middledown workflow. SCIEX technical note, MKT-28341-A.
- Achieving ultrahigh sequence coverage and high-confidence disulfide bond mapping of biotherapeutics using an electronactivated dissociation (EAD)-based middle-down workflow. SCIEX technical note, MKT-28565-A.
- Milos Cejkov et al. (2021) Electron transfer dissociation parameter optimization using design of experiments increases sequence coverage of monoclonal. J. Am. Soc. Mass Spectrom. 32(3): 762-771.
- Jonathan Dhenin et al. (2023) A multiparameter optimization in middle-down analysis of monoclonal antibodies by LC– MS/MS. J. Mass Spectrom. 58(3):e4909.


