Methods
Sample preparation: Plasma extract (NIST, 1g/100mL) is a commercially available lipid mixture that contains diverse lipid species. Available internal standards for retention time determination and for quantitation include the Avanti SPLASH standard mixture and the SCIEX Quantitative Lipid Standards kits.6
LC-MS analysis: The LC separation was performed using a ExionLC system consisting of a binary high pressure mixing gradient pump with degasser, a thermostated autosampler, and a column oven. Data can be acquired using either the QTRAP 6500+ system, equipped with an IonDrive Turbo V ion source, was operated in low mass mode with Analyst® Software or the SCIEX Triple Quad 7500 system equipped with an OptiFlow Pro ion source and operated using SCIEX OS software. Retention times are first roughly determined using either the NIST plasma sample spiked with internal standards or a representative matrix sample and the unscheduled MRM method. Analysis of this preliminary data enables the development of a time-scheduled MRM method leveraging the enhanced features in the Scheduled MRM™ Algorithm. Details on the chromatographic and MS methods can be obtained from the Comprehensive Targeted Method for Global Lipidomics Screening7.
Data processing: All data was processed using SCIEX OS Software using AutoPeak algorithm. Automated computation of the time scheduled MRM methods was performed using the sMRM Pro Builder Template Mx_Lx 1.4.8
MRM approach in lipid identification
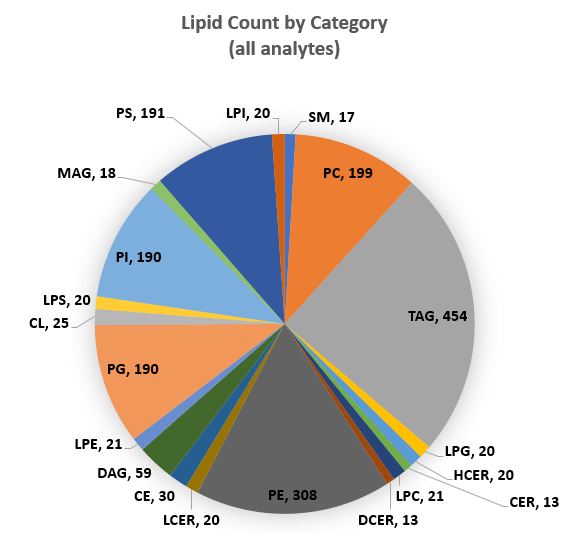
Here a large panel of MRM transitions have been developed to provide a targeted assay for a broad range of lipid species. The targeted list is composed of ~1900 lipid molecular species, including PC/LPC, PE/LPE, PS/LPS, PI/LPI, PG/LPG, MG, DG, TG, CL, CE, CER, DCER, HexCER, LacHCER and SM. Lipid classes currently included in assay provide good coverage of lipids commonly found in mammalian plasma, tissues and cells. Additional lipids can be added to the assay depending on matrix as workflow is easily extensible.
Most lipid classes and categories are acquired in the positive ion mode. However, for the targeting of phospholipids (PL), rather than using the more common approach via the head group loss in positive ion mode, this approach uses negative ion mode via the loss of fatty acid to identify the PL (PC, PE, PS, PI, PG) at the fatty acid composition molecular species level. As shown in Figure 2, the PC 34:2 can only be identified at the class level using the positive ion mode while the specific molecular species can be figured using the negative ion mode. Use of fast polarity switching on the SCIEX systems allows monitoring of both positive and negative MRMs providing a key advantage for this method. This level of molecular species specificity is often key to understanding the specific biology.
Streamlined method development
As the number of lipids covered in this method is large, it is important to use the Scheduled MRM algorithm approach during acquisition of biological dataset for highest quality quantitation results. To streamline the development of this final assay, an Excel based sMRM Pro Builder template8 has been developed to assist the user during the assay refinement steps.
First the user collects MRM data on a representative matrix (with or without internal standards) for the upcoming biological study (3-5 replicates). Results from the first pass unscheduled MRM acquisitions are processed with SCIEX OS software (Figure 3) and then entered into the sMRM Pro Builder and an initial rough approximation of the retention times is determined. Next 5-10 replicate injections are performed on the matrix using a preliminary time scheduled MRM method, ideally a pooled sample from the biological samples to be run. Data analysis here provides information on peak width, RT variance, and lipid signal and the sMRM Pro Builder computes a final optimized Scheduled MRM Acquisition method (Figure 4).
After method construction, the Scheduled MRM algorithm Concurrency tool9 for Analyst software or the sMRM Summary build into SCIEX OS software10 can be used to check the concurrency and hence resulting dwell time for the final assay.
Chromatographic reproducibility
The LC-MRM reproducibility was evaluated by injecting the NIST plasma lipid extract (10 replicates) using the final Scheduled MRM Pro assay. Excellent reproducibility was observed with majority of lipid species showing retention time standard deviations below 0.05 minutes (Figure 5).
Quantitative reproducibility
Using the optimized Scheduled MRM algorithm acquisition method, 5 replicate injections were performed on plasma using the SCIEX Triple Quad 7500 system. Reproducibility of quantitation was assessed by computing the %CV of the peak area replicates for the lipids within each class. Figure 6 (middle) shows the cumulative fraction of lipids in selected classes at each % CV level for all lipids from the optimized method. Nine HexCER lipids were measurable in plasma and HexCER was the 5th most abundant class as measured by sum of peak areas (top) and showed highest reproducibility (middle). Other classes (SM, PC, CER, DCER, HexCER and LPC) were also measured with very good reproducibility with 80% of lipids quantified with 20% CV or better. The TAGs are a large class (454 tested) but by filtering the assay to the 201 TGs measurable in this sample, good reproducibility of this class was obtained (Figure 6 bottom).
Improved results quality with Scheduled MRM Pro algorithm
Advanced method features like Retention Time Tolerance (Flexible Window Width) in Scheduled MRM algorithm allow for additional method optimization to further fine tune an MRM method with many transitions such as this. Using the sMRM Pro Builder template and performing LC-MRM replicates, optimized values can be determined. sMRM Pro Builder template can also automatically reject low intensity peaks (uncertain detection) from final assay to further improve results quality by reducing MRM concurrency. Results obtained using the algorithm after filtering can be compared for 10 replicates of plasma matrix and filtering can be optimized.
Using internal standards for quantitation
It is important to note that quantitation of lipids via global lipid profiling is challenging. In this method, several options are available to quantitate lipid levels in samples. For relative quantitation, a single lipid molecular species for each lipid class is needed. This strategy corrects for extraction efficiency as well as systematic errors; however, this strategy should only be used for comparative analysis between two samples. Using this strategy, it is not correct to report the concentrations of individual lipid molecular species, because the fragmentation efficiency of different molecular species within the same class is not equal and depends on the length and the number(s) of double bonds present in the fatty acid that fragments in the negative ion mode. Using this strategy, it is possible to conclude certain lipids increase or decrease when two different samples are compared. SPLASH mix from Avanti Polar Lipids is a commercially available mixture of lipid internal standards that can be used for this quantitation strategy.
A second strategy involves using multiple labeled internal standards to enable accurate quantitation (quantitative bias <10%); absolute quantitation requires a labeled internal standard for every analyte and is not possible in these experiments. Accurate quantitation can be achieved by assigning a structurally similar (but not necessarily identical) internal standard to each analyte. Recently, a comprehensive mixture of labeled internal standards was developed for this purpose.6 Using these standards, accurate quantitation can be achieved for many but not all of the lipid classes targeted in this method (SM / DG / CE / CER / TG / LPE / LPC / PC / PE / FFA).
The method reported here is flexible to either internal standard strategy through addition of the chosen internal standards to the sample and inclusion in the MRM method development.
Summary
In this study, a targeted global lipid profiling strategy is presented that enables a broad array of different lipids to be quantified at the molecular species level (~1900 molecular species). An optimized chromatography method was developed to minimize isobaric interference through the chromatographic separation of lipid class, which allows for a relatively rapid and specific lipid screening technique. The target list of lipids is comprehensive, covering most major lipid classes and categories, and MRMs were selected to cover lipids containing fatty acids with 12-26 carbons and 0-6 double bonds including odd chain lipids. The method is customizable, so new lipid categories, classes and molecular species can be added.
Results quality of the method has been demonstrated in plasma matrix to highlight both the robustness and the flexibility of the LC-MRM assay.
An easy to use tool (sMRM Pro Builder template) was also developed to streamline the optimization of the targeted MRM assay to maximize results quality when performing assay with very large numbers of time scheduled MRM transitions.

 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge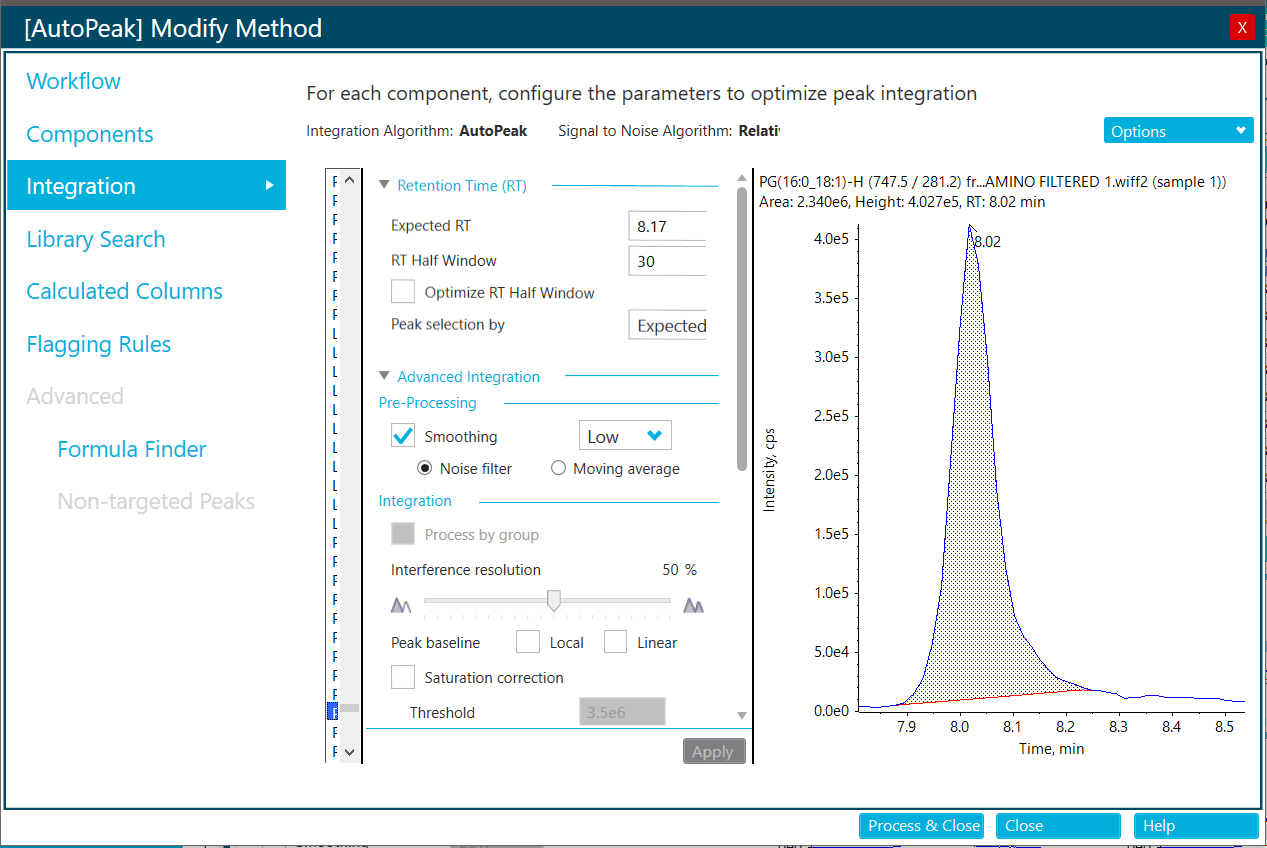 Click to enlarge
Click to enlarge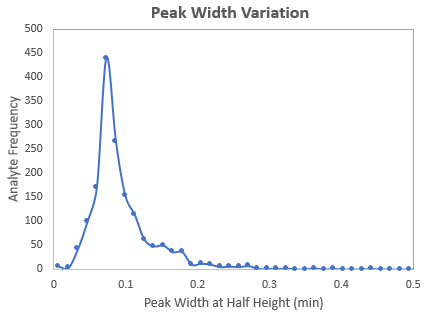 Click to enlarge
Click to enlarge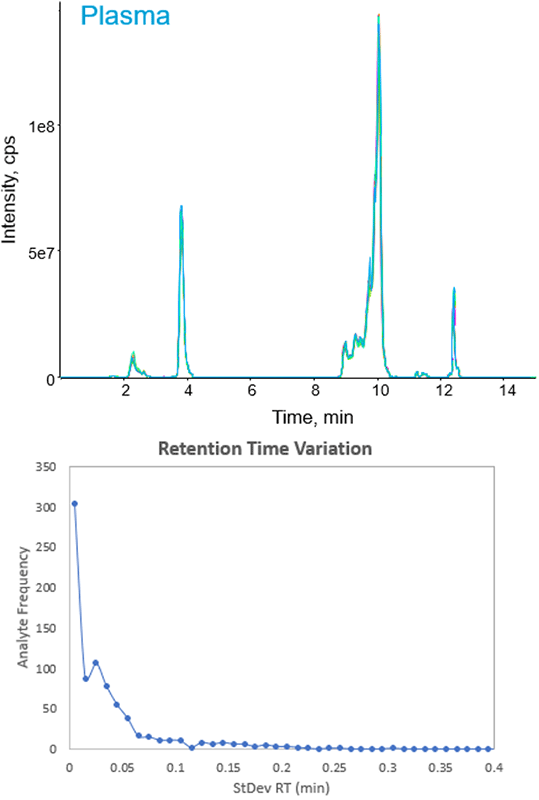 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge