Fast protein identification experiments with Microflow LC – up to 100 samples per day
High speed acquisition with TripleTOF® 6600 System
Nick Morrice1, Zuzana Demianova2, Xiaolei Lv3, Christie Hunter4
1SCIEX, UK, 2SCIEX, Germany, 3SCIEX, China, 4SCIEX, USA
Abstract
Increasingly, microflow liquid chromatography (LC) is being used for the analysis of complex proteomics samples because of higher robustness and faster run times. Here, the use of very fast microflow LC gradients was explored for protein identification experiments, using gradients as short as five minutes. Combined with MS/MS acquisition rates as fast as 66 Hz on the TripleTOF® 6600 System, good protein identification results are achieved providing high sample throughput. Using multiple instruments and multiple complex matrices, it was observed that the number of proteins identified for even the fastest gradients was still over 1000 for multiple complex matrices from 1 µg protein load.

Introduction
One of the primary challenges to producing statistically significant results for proteomics has been the long times required for data acquisition using nanoflow chromatography. Even relatively simple protein identification experiments require up to one hour of instrument time per sample due to long sample injection, column washing and equilibration times. Microflow liquid chromatography (LC) provides more reproducible separations and more robust electrospray, and is being used increasingly for the analysis of complex proteomics samples. Additionally with higher flow rates, less time is required to load, wash and equilibrate the trap and column.
Data presented here demonstrates the high-throughput workflows now possible for data dependent acquisition (DDA) using microflow liquid chromatography gradients as short as five minutes (total run time <15mins). The combination of microflow LC with the high MS/MS acquisition rates of the TripleTOF® 6600 System enables high-throughput yet comprehensive analysis for proteomics samples, at rates approaching 100 samples per day.
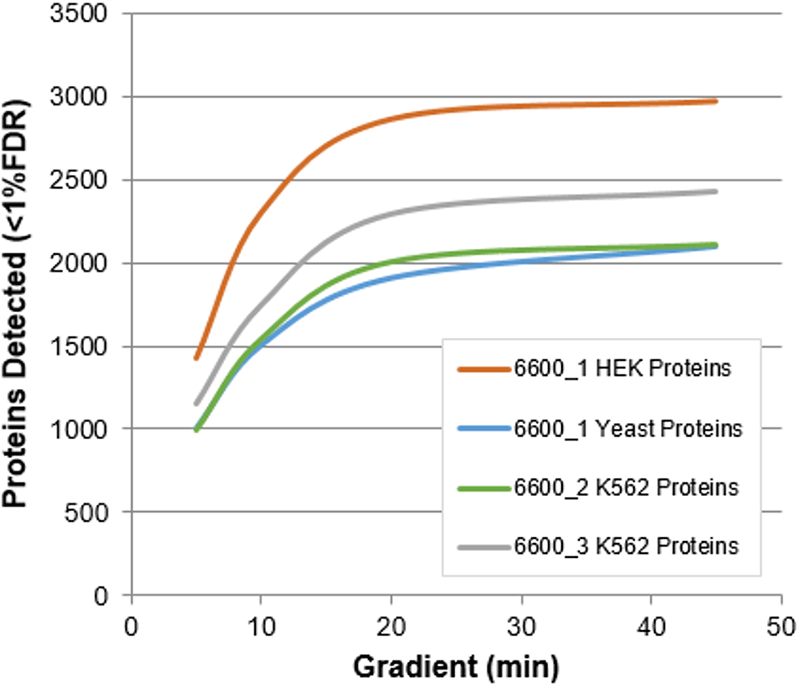
Using multiple instruments and multiple complex matrices, it was observed that the number of proteins identified for even the fastest gradients was still over 1000 for multiple complex matrices from 1 µg protein load (Figure 1).
Figure 1. Impact of gradient length on protein identification Rates. Using microflow LC, a series of different gradient lengths were explored and the number of proteins identified were determined from an on-column load of 1 µg total protein. Even using the very fast gradients, 1000-2000s proteins were confidently identified. Data was repeated on 3 different instruments with 3 different matrices to test the robustness of the methods and similar results were observed.
Key feature of fast microflow LC workflow
- TripleTOF 6600 System with the Accelerator TOF™ Analyzer can easily match the fastest LC workflows.
- High resolution MS/MS can be acquired at rates of up to 100 MS/MS per second.
- Optimized collision energy settings3 ensures high quality MS/MS for robust peptide identification
- The versatile NanoLC™ 425 System enables the researcher to span the flow regimes from nanoflow to high microflow easily.
- Dual gradient system with interchangeable flow modules for maximum workflow flexibility
- Minimal delay volume enables fast formation of microflow gradients.
- Powerful algorithms in ProteinPilot™ Software ensures high numbers of spectra are identified.
- Paragon™ Algorithm allows hundreds of modifications and substitutions to be searched for simultaneously using its unique hybrid algorithm, combining database searching with a novel sequence tag search method.4
- Pro Group™ Algorithm applies rigorous protein inference for reporting reliable, defensible protein identifications.
Methods
Sample preparation: Cell lysates (HEK, K562 and Yeast) were digested with trypsin using standard protocols. Sample loading of 0.5 – 2 µg of total protein were used for each injection.
Chromatography: A NanoLC™ 425 System plumbed for microflow chromatography was used (5 µL/min) and operated in trap/elute mode. Column temperature was controlled at 30°C. Gradients of 5, 10, 20, or 45 minutes were tested (Table 1). More information on LC configuration can be found in the SWATH Performance Kit SOP.1
Mass spectrometry: All data was acquired using a TripleTOF® 6600 System with the Turbo V™ Source equipped with the 25 µm hybrid electrodes for microflow LC. IDA data were collected using a variety of acquisition strategies. The TOF MS scan was 150 msec and the number of MS/MS per cycle and accumulation time was varied. Other method parameters were varied as described for Figure 3.
Data processing: Data dependent acquisition (DDA) data was processed using ProteinPilot Software 5.04 and the FDR reports were evaluated using the FDR Comparison Template2 provided with the software.
Table 1. Gradient profiles used for the protein identification comparisons.
Figure 2. High gradient reproducibility. Even for that fastest gradients, very reproducible ion chromatograms were observed. TOF MS TICs for the 5 and 10 minute gradients are shown.
Optimization of protein ID settings
A series of optimization experiments were performed to test the best acquisition conditions for running data dependent experiments under accelerated chromatographic conditions. As faster gradients will produce taller sharper LC peaks, parameters such as intensity threshold for MS/MS trigger, number of MS/MS per cycle and accumulation time (total cycle time), exclusion time after MS/MS would need to be adjusted.
Figure 3 shows a few examples of the parameter optimization ramps that were performed. Increasing the number of MS/MS per cycle provided significant improvements in the number of IDs. The accumulation time per MS/MS was also decreased to maintain cycle time and it was found that very fast acquisition rates (accumulation times down to 15 msec) provided results improvements. There was little dependence on intensity threshold, perhaps due to the overwhelming # of precursors available in every cycle, however the threshold was increased to 500 or 1000 for the very fast MS/MS experiments to ensure data was acquired at points higher up on the LC peak. At these very fast accumulation times (15 msec), ~70% of the acquired spectra yielded peptide identifications, indicating the high spectral quality.
Optimized collision energy values for peptides3 were used, and for the very low accumulation times, collision energy spread (CES – short CE ramp around optimized values) was turned off. This was found to have minimal impact on results quality.
Figure 2. High gradient reproducibility. Even for that fastest gradients, very reproducible ion chromatograms were observed. TOF MS TICs for the 5 and 10 minute gradients are shown.
Figure 3. Parameter optimization ramps for fast gradients. A variety of parameters were varied (table) and impact on protein identification rates were assessed (plot). The results for the HEK cell lysate using the 5 minute gradient is shown, # of peptides (open circles) and proteins (closed circles) plotted for instrument 1. Assessment of the MS/MS quality showed that 50-60% of MS/MS acquired still had scores >12 for these very fast acquisition rates.
Evaluating protein loading
A few different protein loads were tested using these faster gradients to understand the impact on protein identification rates. Figure 4 shows the results from the 10 and 20 minute gradients across 3 different protein loadings for K562 digest on one of the TripleTOF Systems tested. The number of peptides identified increased by about 13% when the load was increased from 0.5 to 2 µg resulting in an 8% increase in protein IDs.
Figure 4. Impact of protein load on ID rates. Results from the 10 minute (blue) and 20 minute (green) are shown for 3 different proteins loads (0.5,1, 2 µg) for the K562 digest on instrument 3. The protein numbers are shown as solid bars and the peptide numbers are shown as the line graphs.
Impact of gradient length on results
Using a protein load of 1 µg, the acquisition conditions determined best for each gradient length, a final dataset was collected to show the ID trends (Figure 1). Here, the numbers shown were generated using the acquisition method with 90 MS/MS per cycle and 15 msec accumulation time, providing 66 Hz acquisition rates for the 5, 10 and 20 min gradient. 60 MS/MS x 25msec was used for the 45 minute gradient. The number of proteins and peptides identified by ProteinPilot Software at <1% global FDR5 were reported.
Very similar numbers of peptides were detected between the 3 instruments on the different matrices (Figure 5). Very little decrease is seen as the gradient was shortened from 45 to 20 mins. At 10 mins, about 9000 peptides are observed at < 1% global FDR. Even for the 5 min gradient, 5000 peptides were identified.
The relative numbers of proteins identified at the various gradients lengths were plotted as well to provide researchers a guidance when selecting the right LC strategy for a study (Figure 6). Comparing to the protein numbers obtained with the typical 45 min gradient used for microflow, small decreases in protein numbers were observed at shorter run times. However, even with the shortest gradients of 5 mins, 50% of proteins were still identified, suggesting this is a compelling strategy for fast interrogation of proteomics samples.
Figure 5. Peptide identification results for fast microflow gradients. The # of peptides identified from 1 µg protein load are show for 3 different instruments on 3 different matrices.
Figure 6. Selecting the best chromatographic strategy. Plotting the proteins identified relative to the numbers obtained on the typical longer 45 min gradient, the resulting decrease in the protein ID numbers is surprising low. Using a 10 minute gradient, about 70% of the proteins are still identified, and even at 5 mins, 50% of proteins were still confidently identified relative to the longer gradient.
Conclusions
Microflow LC is proving to be a robust and useful tool for advancing and accelerating proteomics research. Here the use of very fast LC gradients was explored, using microflow LC gradients as short as five minutes. Combined with MS/MS acquisition rates as fast as 66 Hz on the TripleTOF 6600 System, good protein identification results are achieved providing high sample throughput.
Protein ID rates were nearly as good for 20 min as for a 45 min gradient for both peptides and proteins using microflow LC. ID rates drop using faster gradients but are still quite high when the 10 and 5 minute gradients are used (ie. for 10 min, ID ~50 and 75% respectively of peptides and proteins found at 45 mins).
When more rapid protein ID results are required or when less complex samples are to be analyzed, the fast microflow LC approach with TripleTOF Systems provides a high-throughput robust solution.
References
- SWATH Performance Kit, SCIEX product page.
- FDR Comparison Template for Protein ID. SCIEX community post RUO-MKT-18-10968-A.
- Optimized Collision Energy Curves for TripleTOF Systems. SCIEX community post RUO-MKT-11-2649-A.
- ProteinPilot™ Software overview. SCIEX technical Note RUO-MKT-02-1777-B.
- Global vs Local FDR Rates. SCIEX community post RUO-MKT-18-8310-A.
- Accelerating SWATH® Acquisition for Protein Quantitation – Up to 100 Samples per Day. SCIEX technical note RUO-MKT-02-8432-A.
 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge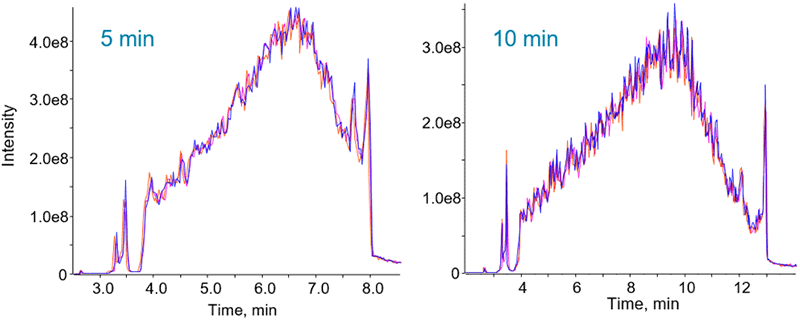 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge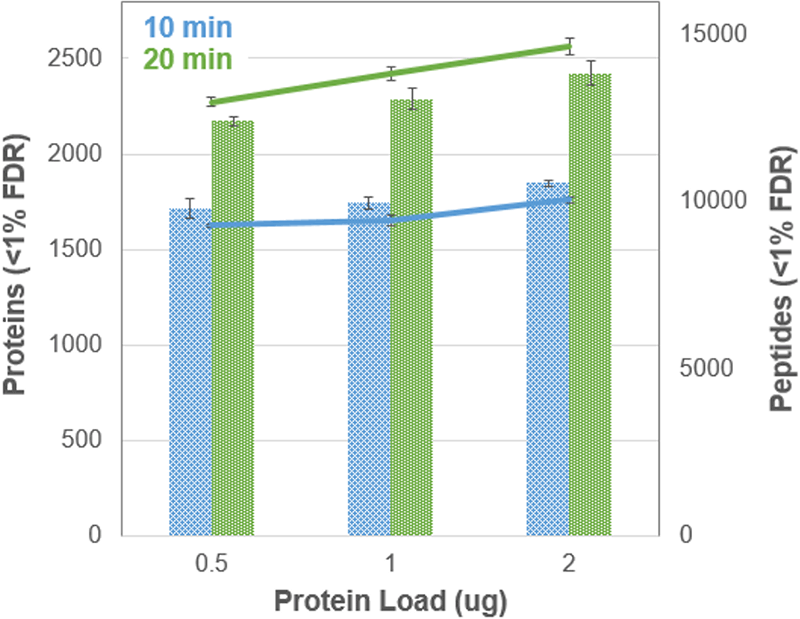 Click to enlarge
Click to enlarge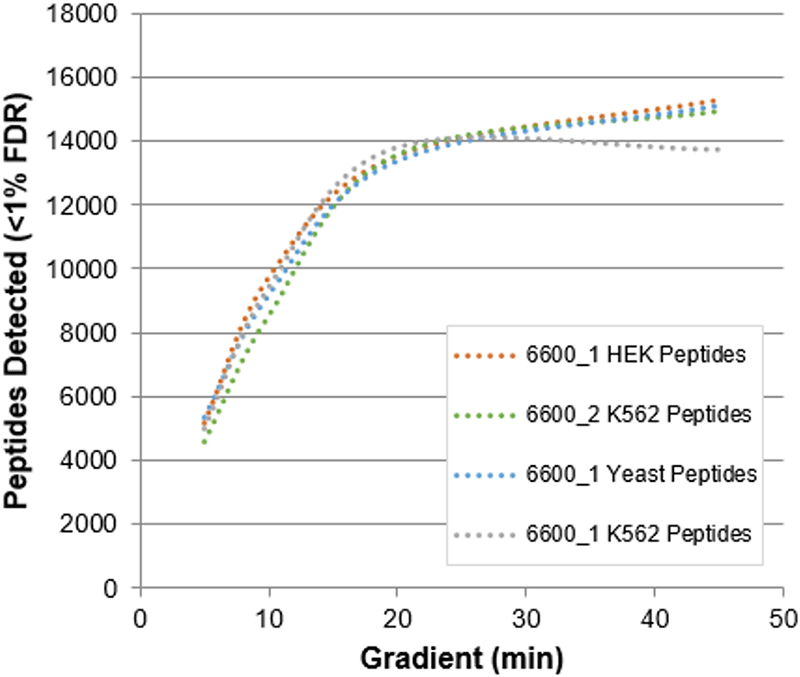 Click to enlarge
Click to enlarge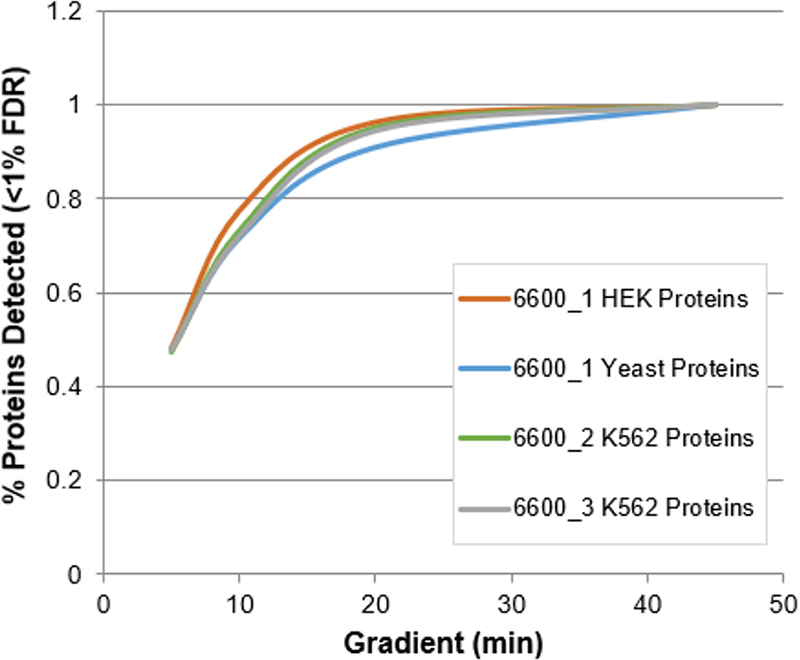 Click to enlarge
Click to enlarge