Accelerating SWATH® Acquisition for protein quantitation – up to 100 samples per day
Microflow LC with TripleTOF® 6600 System
Christie Hunter1, Nick Morrice2, Zuzana Demianova3
1SCIEX, USA, 2SCIEX, UK, 3SCIEX, Germany
Abstract
The combination of microflow LC with SWATH Acquisition for large scale quantitative proteomics studies is becoming increasingly more widespread, due to the improved robustness and throughput obtained relative to the traditional nanoflow LC approach. Here, the impact of shorter gradient lengths on the number of proteins that can be quantified by SWATH Acquisition using the TripleTOF 6600 system was investigated. Due to the very fast MS/MS acquisition rates possible on TripleTOF systems, high numbers of peptides/proteins can be quantified with very fast gradients. For example, with the Pan Human Library on a 1ug load of HEK lysate, ~2100 and ~3400 proteins were quantified using SWATH with the 5 and 10 min gradients respectively.

Introduction
The combination of microflow LC with SWATH Acquisition for large scale quantitative proteomics studies is becoming increasingly more widespread, due to the improved robustness and throughput obtained relative to the traditional nanoflow LC approach. High quality quantitative datasets have been generated using a standard 1 hour gradient, demonstrating large numbers of proteins quantified routinely.1
Previously, the impact of gradient length on protein identification results using data dependent acquisition (DDA) was explored.2 Here, an exploration into the impact of gradient length on proteins quantified using data independent acquisition (DIA) was undertaken to provide researchers expanded workflow options with microflow SWATH Acquisition. Using microflow liquid chromatography gradients as short as five minutes (total run time <15mins), SWATH Acquisition parameters were first optimized then used on multiple TripleTOF® 6600 Systems to study impact of shortened separations on number of proteins quantified.
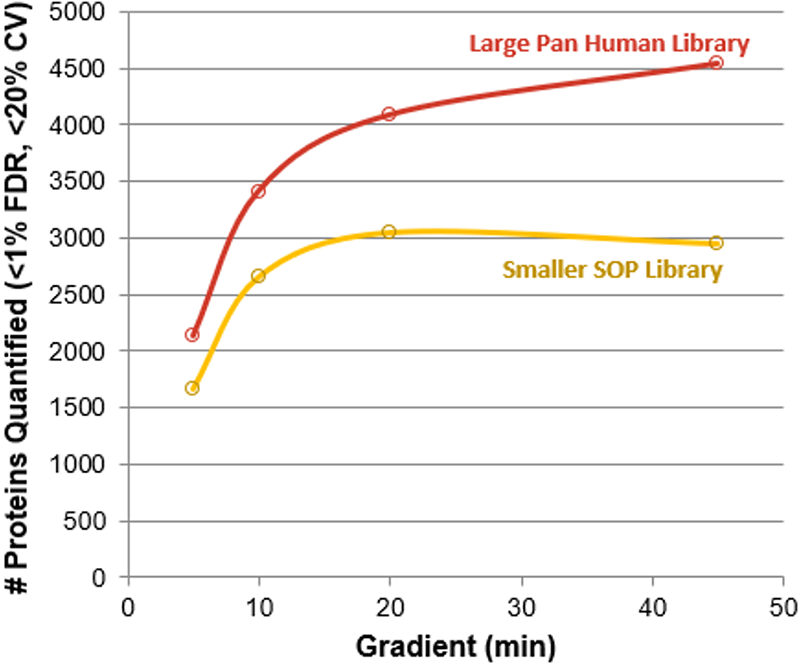
Testing on multiple instruments with different complex matrices, it was observed that the number of proteins quantified using SWATH Acquisition for the fastest gradients was over 1300 proteins for multiple complex matrices from 1 µg protein load (Figure 4). When combined with larger ion libraries, over 2100 proteins were identified and quantified in a 5 min gradient from a complex HEK digest (1 µg, Figure 1). The combination of microflow LC with the high MS/MS acquisition rates of the TripleTOF® 6600 System enables high-throughput yet comprehensive analysis for proteomics samples, at rates approaching 100 samples per day.
Figure 1. Impact of gradient length on SWATH Acquisition quantitation results. The replicate data collected at each gradient length was processed with both the smaller SOP library (yellow) and the larger Pan Human Library (red). This was done using the HEK lysate (1 µg load) on instrument 3. In each case, significant gains in proteins quantified was observed at each gradient length with the larger library, on average more than 30% more proteins.
Key features for fast quantitation
- TripleTOF 6600 System with the Accelerator TOF™ Analyzer can easily match the fastest LC workflows.
- High speed MS/MS enables the use of 100 variable sized Q1 window isolation3 for SWATH Acquisition, for improved data quality through increased specificity, even at fastest gradient speeds.
- Optimized collision energy settings5 ensures high quality MS/MS for robust peptide identification.
- Versatile NanoLC™ 425 System enables the researcher to use multiple flow regimes, from nanoflow to high microflow easily, on a single LC system.
- Dual gradient system with interchangeable flow modules for maximum workflow flexibility.
- Minimal delay volume enables fast formation of microflow gradients.
Methods
Sample preparation: Cell lysates (HEK, K562 and Yeast) were digested with trypsin using standard protocols. Sample loading of 1 µg of total protein were used for each injection.
Chromatography: A NanoLC™ 425 System plumbed for microflow chromatography (5 µL/min) was used and operated in trap/elute mode. Column temperature was controlled at 30°C. Gradients of 5, 10, 20, or 45 minutes were tested (Table 1). More information on LC configuration can be found in the SWATH Performance Kit SOP.4 Note the wash and equilibration steps were not optimized, further reductions in run time may be achievable.
Mass spectrometry: All data was acquired using a TripleTOF® 6600 System with the Turbo V™ Source equipped with the 25 µm hybrid electrodes for microflow LC. SWATH Acquisition data were collected using a variety of acquisition strategies using variable Q1 windows. The TOF MS scan was 150 msec and the # of Q1 windows/cycle and accumulation time was varied. Use of collision energy spread (CES - short CE ramp around optimized values) was investigated in combination with optimized collision energies5.
Data processing: Data independent acquisition (DIA) data was processed using SWATH Acquisition microapp in PeakView® Software 2.2. Results were evaluated using the SWATH Replicates Template.6 Two ion libraries were used for processing, the SWATH Performance kit library4 and the Pan Human Library.8
Table 1. Gradient profiles used for the protein quantitation comparisons.
Optimization of SWATH Acquisition methods
A series of optimization experiments were performed to test the best acquisition conditions for running DIA experiments under accelerated chromatographic conditions. As faster gradients produce taller sharper LC peaks, cycle times need to be considered for best quantitation results, therefore the number of Q1 windows per cycle and accumulation time were adjusted.
Figure 2 shows a few examples of the parameter optimization ramps that were performed. Increasing the number of MS/MS per cycle provided significant improvements in the number of IDs. The accumulation time per MS/MS was also decreased to maintain cycle time and it was found that very fast acquisition rates (accumulation times down to 15 msec) provided results improvements. This optimization was performed for each gradient length. The XIC window width with for the shorter gradients was also varied (1 – 4 min for the 10 min gradient) and there was minimal impact on proteins quantified (data not shown).
Optimized collision energy values for peptides5 were used, and for the very low accumulation times, CES) was turned off. This was found to have minimal impact on results quality (Table 2).
Figure 2. Parameter optimization for fast gradients. A variety of parameters were varied and the impact on # of proteins (blue) and peptides (orange) quantified was assessed. The results for the HEK cell lysate (1 µg) using the 10 minute gradient is shown for instrument 1. Increasing the # of Q1 windows used per cycle with faster accumulation times provided increased specificity for these compressed gradients and thus provided gains in quantified proteins and peptides.
Table 2. Evaluating the impact of collision energy spread (CES). Minimal impact on the numbers of proteins / peptides quantified by SWATH Acquisition was observed when collision energy spread (CES) was on (5 V) vs off (0 V).
Impact of using larger libraries vs gradient length
To understand if shorter gradients have an impact on protein quantitation results when using larger libraries, the Pan Human Library8 was used to process the HEK data obtained off instrument 3 (with both PepCalMix and iRT peptides dosed in). The same data was also processed with the SWATH Performance Kit SOP library4 for comparison. At each gradient length, significant gains in proteins quantified was observed, on average over 30% more proteins were quantified with the larger library (Figure 3). Even larger gains were observed in the number of peptides quantified using the larger Pan Human Library, on average over 75% more peptides were quantified with the larger library at each gradient length (data not shown).
Figure 3. Gains observed when using larger Pan Human Library (PHL) across gradients. This plot represents the same data as in Figure 1 but plotted in terms of % increase for proteins (blue) and peptides (orange) quantified. The larger library provided on average more than 30% more proteins and 70% more peptides across the different gradient lengths.
Impact of proteins quantified vs gradient length
Using a protein load of 1 µg and the optimized SWATH Acquisition methods (Table 3), a final dataset was collected across 3 different TripleTOF 6600 Systems on different matrices (Figure 4). For the faster gradients, accumulation times of 15 msec were used for MS/MS, providing 66 Hz acquisition rates. This high speed MS/MS acquisition allowed a maximal # of Q1 windows to be used to provide higher specificity as the separation was compressed.
Similar numbers of peptides were detected between the 3 instruments on the different matrices (Figure 4, bottom) and typically only a 50% loss in # of peptides quantified between 45 min and 5 min gradient was observed. As expected, the number of proteins quantified across matrices varied (Figure 4, top), however similar to what was observed with the number of peptides quantified, 50% of proteins were still quantified using the 5 min gradient, suggesting this is a compelling strategy for fast interrogation of proteomics samples.
Table 3. Optimized SWATH Acquisition conditions. After testing various acquisition conditions on the multiple TripleTOF Systems, these final conditions were used to collect the final datasets.
Figure 4. Proteins and peptides quantified from SWATH Acquisition Data. Results from the various gradient lengths for the different matrices on the 3 different TripleTOF 6600 Systems tested are plotted, proteins (top) and peptides (bottom). Protein load for all data is 1 µg.
Conclusions
Understanding the impact of gradient length on proteins quantified will provide researchers workflow options depending on the sample complexity and the size of the study (# of samples to be run) when performing microflow SWATH Acquisition studies. Here, the impact of very fast LC gradients on proteins and peptides quantified reproducibly and with high confidence was explored on multiple systems.
- Multiple instruments and multiple complex matrices were tested to obtain a robust understanding of the tradeoffs between running shortened gradients with microflow SWATH Acquisition.
- The very fast MS/MS acquisition rates (up to 66 Hz) of the TripleTOF 6600 System enables use of higher numbers of Q1 SWATH Acquisition windows to be used even with the fastest gradients, providing specificity and robust quantitation.
- The impact of library size was also tested on the accelerated gradients and provided improved results as expected. With the Pan Human Library on a 1ug load of HEK lysate, ~2100 and ~3400 proteins were quantified with the 5 and 10 min gradients respectively.
References
- Microflow SWATH® Acquisition for industrialized quantitative proteomics. SCIEX technical notes RUO-MKT-02-3637-A.
- Fast protein identification experiments with microflow LC – up to 100 samples per day. SCIEX technical notes RUO-MKT-02-8312-A.
- Evolution of SWATH® Acquisition Provides Large Gains in Quantified Proteins. SCIEX technical notes RUO-MKT-02-5772-A.
- SWATH Performance Kit, SCIEX product page.
- Optimized Collision Energy Curves for TripleTOF Systems. SCIEX community post RUO-MKT-11-2649-A.
- Download the SWATH Replicates Template.
- Extending Depth of Coverage with SWATH® Acquisition Using Deeper Ion Libraries. SCIEX technical notes RUO-MKT-02-3247-A.
- Rosenberger G et al. (2014) A repository of assays to quantify 10,000 human proteins by SWATH-MS. Scientific Data, 1, 140031.
 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge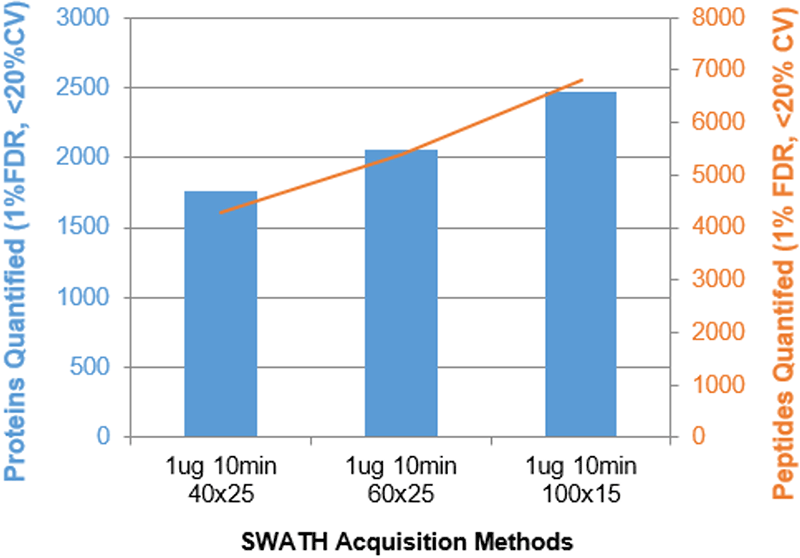 Click to enlarge
Click to enlarge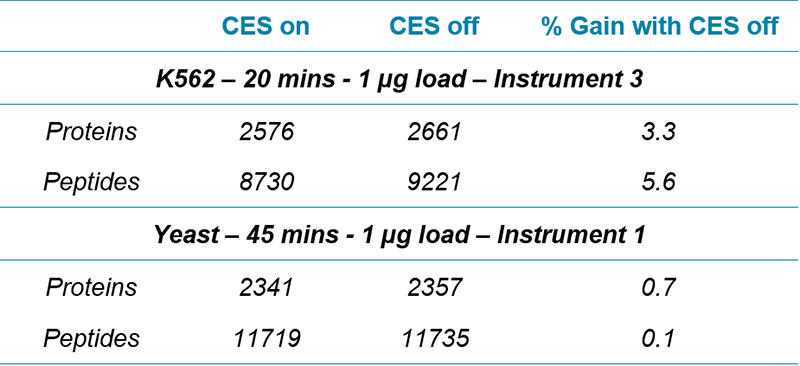 Click to enlarge
Click to enlarge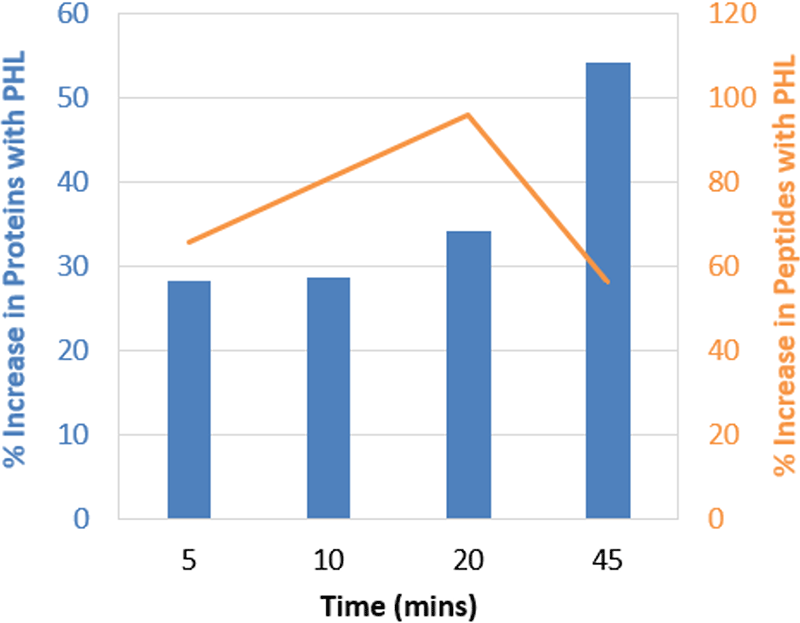 Click to enlarge
Click to enlarge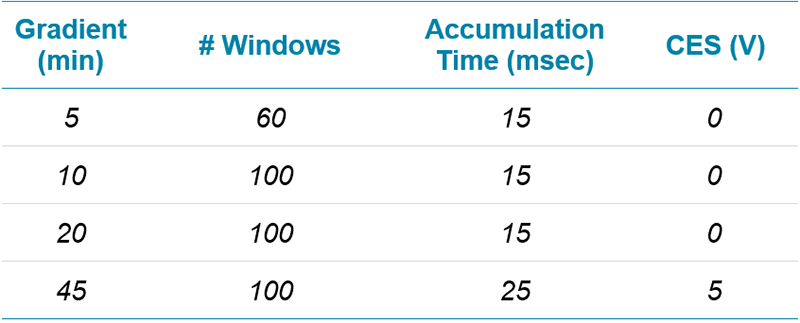 Click to enlarge
Click to enlarge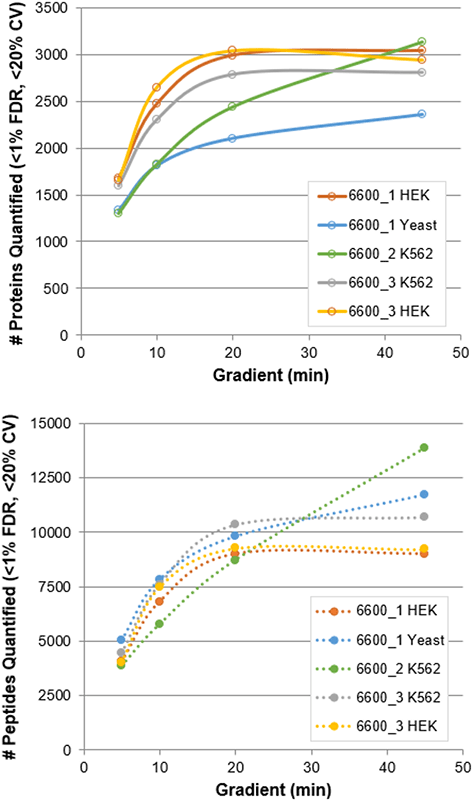 Click to enlarge
Click to enlarge