Using differential mobility spectrometry to separate and localize sites of post-translational modifications on peptides
Using SelexION Differential Mobility device on QTRAP 5500 system
Ji Luo1, Jianlan Yu1, Wenhai Jin1, Rong Zeng2, Yongming Xie1, Christie Hunter3
1SCIEX, China; 2SIBS, China; 3SCIEX, USA
Abstract
Differential mobility spectrometry (DMS), known as the SelexION device, can be used to separate peptides based on different chemical properties and to separate isobaric or isomeric compounds. Here, peptides with different modification sites were analyzed using SelexION Technology on the QTRAP 5500 system.

Introduction
Characterization of the post-translational modification sites (PTMs) on proteins and peptides is a challenging task. Mass spectrometry can provide a good solution for characterization because of the high quality MS/MS spectra that can be obtained for identification and site localization. However, isomeric peptides with PTMs on different residues of the same peptide sequence can be hard to distinguish by MS/MS, especially if they co-elute during chromatographic separation. This can lead to ambiguous assignment of the modification sites, even when using high resolution MS. Likewise, trimethylation and acetylation modifications are nearly isobaric and have very similar hydrophobicity, which makes peptides possessing these modifications very difficult to distinguish.
Differential mobility spectrometry (DMS) on QTRAP Systems, known as SelexION Technology, can be used to separate peptides based on different chemical properties and can be functional in the separation of isobaric or isomeric compounds. In this study, peptides with different modification sites or types were analyzed using SelexION Technology on the QTRAP 5500 System.
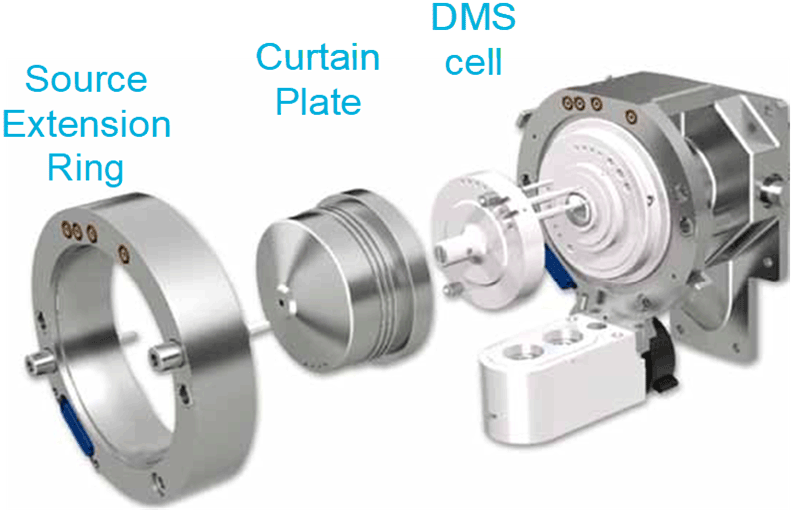
Figure 1. High selectivity quantification using SelexION Technology on the QTRAP 5500 System. The SelexION Technology1 is an easy to install differential mobility spectrometry (DMS) device that can be installed on a QTRAP 5500, 6500 or 6500+ LC-MS/MS System and is used to provide additional selectivity to any quantitative experiment. An asymmetric waveform alternates between high field and low field with an associated separation voltage. In this environment ions will have a net drift towards one of the plates based on their difference in mobility between the high and low fields. A separation voltage (SV) is applied as the filtering voltage and the compensation voltage (CoV) is applied as the restoring voltage, which can be tuned for the compound of interest. Other co-eluting nearly isobaric species that tune with different compensation voltages will be filtered away.
SelexION Technology for orthogonal separation of peptides
- SelexION Technology is a planar differential mobility device (DMS) that separates peptides based on differences in their chemical properties, prior to entering the instrument orifice, thus providing an orthogonal level of selectivity.1,2
- SelexION Technology has been successfully applied in studies of small molecules, including the separation of mixtures of isobaric compounds and isomers3
- In this study, SelexION Technology was utilized for the separation and characterization of isomeric peptides with post-translational modifications on different amino acid residues.
- Coupled with QTRAP System technology, the separation and characterization of isobaric peptides can be achieved.
Methods
Sample preparation: A total of ten synthetic peptides with various modifications were obtained from GL Biochem (Shanghai, China). Peptides were dissolved in 0.1% formic acid at a final concentration of 1 pmol/µL. Peptides with the same amino acid sequence but different modification sites / types were mixed as one sample, for a total of 5 different samples. The sequences of the peptide isoforms studied are shown in Table 1.
HPLC conditions: Samples were analyzed using the microflow LC system. The peptides were directly loaded onto a 0.3 x 50 mm column (C18 AQ, 120 Ǻ, 3 µm, SCIEX) and an elution gradient of 5-40% acetonitrile (0.1% formic acid) over 5 min was used with a flow rate at 10 µL/min.
MS/MS conditions: A QTRAP 5500 System (SCIEX) equipped with Turbo V™ Ion Source and 25 µm I.D. hybrid electrode was used. The instrument was equipped with SelexION Device for differential mobility separations. Compensation voltages (CoV) for all the peptides and isoforms were determined using on-column CoV mapping (Figure 2) and the MIDAS™ Workflow was used to collect full scan MS/MS to confirm the various peptide isoforms. The separation voltage (SV) was fixed at 4000 V, DMS temperature was set to low and no chemical modifier was introduced.
Data processing: Results were analyzed using Bio Tool Kit in PeakView® Software.
Table 1. Post-translation modifications and peptide sequences. A set of standard peptides were generated that possessed PTMs of interest on different locations within the same peptide sequence.
Exploration of separation of post-translational modifications
The peptide mixtures were analyzed using a very short LC gradient (5 min) to simulate the co-elution of the peptides. As the peptides are isobaric, the orthogonal separation strategy provided by the SelexION Device is necessary to separate the peptides using this short gradient.
As a first step in CoV tuning, the peptides were monitored using Multiple Ion Monitoring (MIM) mode, where both Q1 and Q3 were set to the precursor m/z for each peptide, and a series of different CoVs were explored (Figure 2). The LC chromatogram for the two isobaric phosphorylated peptides shows that the peptides could not be chromatographically resolved from each other. However, two distinct CoV optimums were observed, indicating successful separation of the isobaric phosphopeptides by SelexION Technology (Figure 2, top). Using the CoV optimum for each form, full scan MS/MS were triggered and the modification sites assigned without interferences from the other peptide form (Figure 2, bottom).
Figure 2. On-column tuning of compensation voltage (CoV) for phosphorylated peptides. Using Multiple Ion Monitoring (MIM) to specifically target the two isobaric forms of the peptide of interest (FDpSDEDDSTSRF and FDSDEDDpSTSRF) (top), a series of CoV values can be stepped through in an LC-MS time scale to determine the optimum. In this case, two optimal CoV values are observed, one for each peptide isoform. Full scan MS/MS is triggered at the different CoV optimums to confirm the identity of each phosphorylated peptide isoform (bottom).
Phosphorylation
Phosphorylation is one of the key post-translational modifications in cell signaling, and mass spectrometry has been used to identify and confirm large numbers of phosphorylation sites. However, proteins can be phosphorylated at different sites for distinct functions, thereby producing many localization variants of phosphorylated proteins and peptides in biological samples. These modified peptides have the same sequence but the phosphate modification is on different amino acid residues, making them isobaric and quite similar in terms of chromatographic separation. As a result, the MS/MS spectra acquired can be a mixed spectrum where the location of the phosphorylation site is hard to unambiguously confirm.
The ability of SelexION Technology to separate isobaric phosphopeptides was explored using localization variants of phosphorylated peptides. The on-column tuning procedure as described in Figure 2 was used to study a set of modified peptides and obtain MS/MS spectra for characterization. The phosphorylation site on each peptide isoform can be correctly assigned by either manual interpretation or a database search.
Singly phosphorylated peptides were analyzed and data is shown in Figure 2 as described above. The localization variants of di-phosphorylated peptides were also successfully separated by DMS (Figure 3), resulting in clear identification of the different phosphorylation sites for the co-eluting peptides.
Figure 3. Separation of different phosphorylated forms enables definitive characterization. Similar to the analysis procedures in Figure 2, the optimal CoV values were first determined for the two isoforms of di-phosphorylated peptides, and their MS/MS were then acquired for the localization of modification sites.
Methylation and Acetylation
Methylation and acetylation play important roles in many biological activities, especially serving as part of the “histone code” that controls many nuclear processes. There are many different acetylation and methylation modification patterns possible on histone peptides which can be challenging to clearly identify and distinguish.
To demonstrate the potential of DMS in the separation of modified histones, peptides with different acetylation and methylation sites from the histone H3.1 protein were used. Figures 4 and 5 show that SelexION Technology can successfully separate the localization variants of methylated and acetylated peptides.
Figure 4. Separation of methylated peptide isoforms. Isoforms of methylated peptides, where the methyl modification is located on different lysine residues, are shown to co-elute here but can be successfully separated by SelexION Technology because they have different CoV values.
Figure 5. Separation of acetylated peptide isoforms. These co-eluting acetylated peptides with different acetylated lysine residues can also be successfully separated by SelexION Technology due to having different optimal CoVs.
Acetylation and Trimethylation
Acetylation and trimethylation are common modifications occurring on histones that are challenging to distinguish from each other because they are nearly isobaric in mass (Figure 6, bottom). The compensation voltages for a peptide with either acetyl or trimethyl on the N-terminal lysine were measured, and the peptides well separated using DMS, thus allowing for distinct identification of each peptide form.
Figure 6. Separating acetylation from trimethylation during histone peptide characterization. Typically only high resolution instruments have been used to distinguish acetylation from trimethylation as they are nearly isobaric in mass (bottom). Distinct CoV optimums were observed for the acetylated and trimethylated histone peptides (top), allowing them to be successfully separated using DMS.
Conclusions
The study of post-translational modifications on proteins and their role in biology is an important area of study, and tools that can help identify and characterize these sites with more confidence are critical.
- Differential Mobility Spectrometry (DMS) provides an orthogonal level of selectivity by separating components based on their chemical properties and mobility.
- Isobaric co-eluting peptide isoforms where specific modifications are found on different sites of the peptide may be separated using DMS. More peptides will need to be explored to fully understand the utility of this strategy for the separation isobaric peptides with differing modification locations.
- Full scan MS/MS can be obtained at the unique compensation voltage for each peptide isoform to achieve a clean spectrum and confident modification site localization.
- Modifications studied in this work include phosphorylation, acetylation, methylation and trimethylation and demonstrate the potential of SelexION Technology for use in the separation of isobaric biological modifications.
References
- Schneider et al, (2010) Planar Differential Mobility Spectrometer as a Pre-Filter for Atmospheric Pressure Ionization mass Spectrometry, Int. J. Mass Spectrometry, 298 (1-3), 45-54.
- SelexION® Technology: the solution to selectivity challenges in quantitative analysis. SCIEX technical note RUO-MKT-02-3251-A.
- Schneider et al., (2010) Chemical Effects in the Separation Process of a Differential Mobility/Mass Spectrometer System, Analytical Chemistry, 82, 1867-1880.
 Click to enlarge
Click to enlarge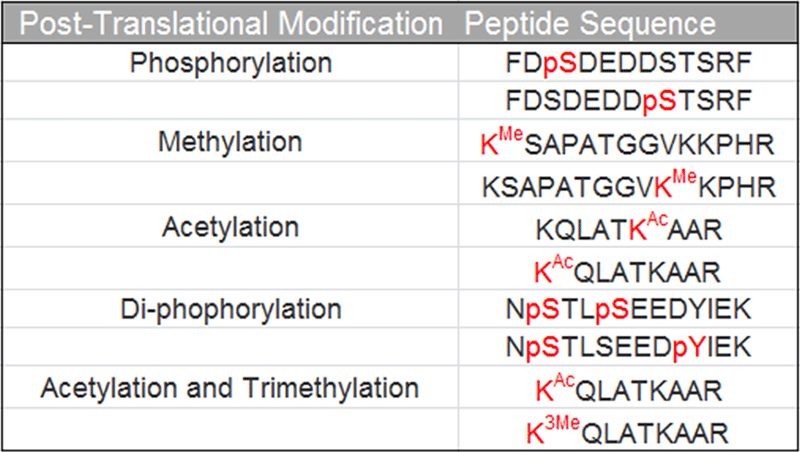 Click to enlarge
Click to enlarge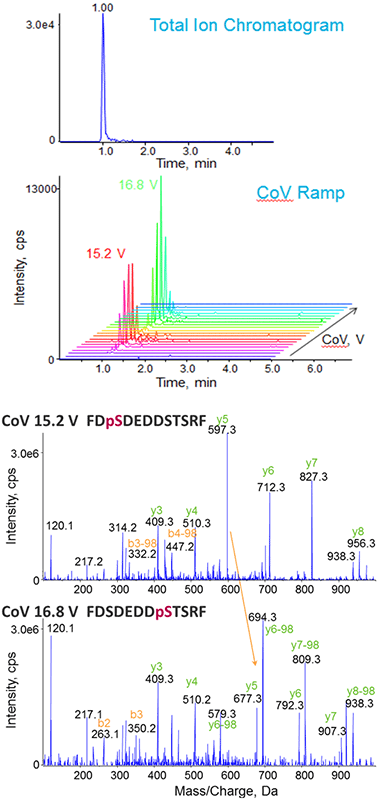 Click to enlarge
Click to enlarge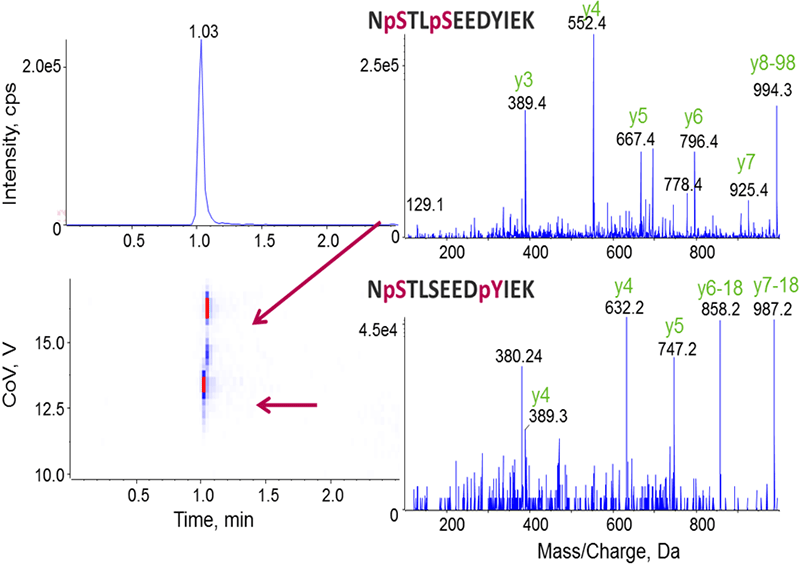 Click to enlarge
Click to enlarge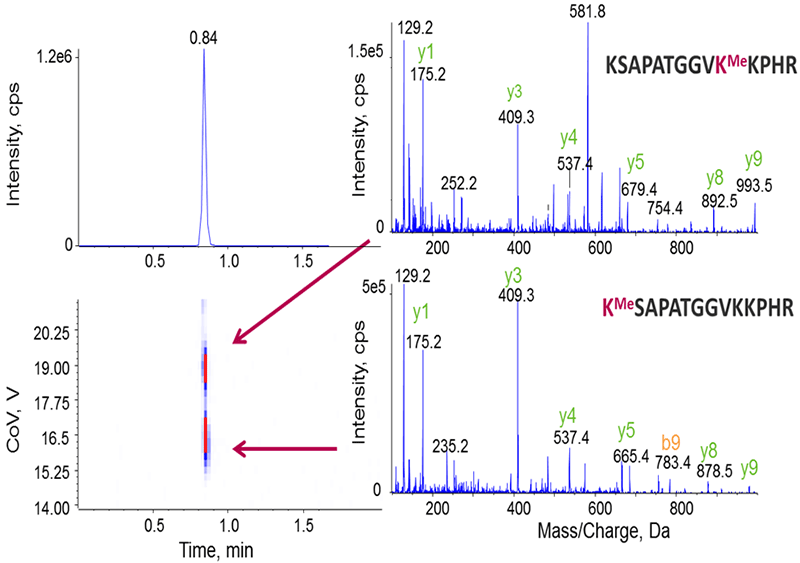 Click to enlarge
Click to enlarge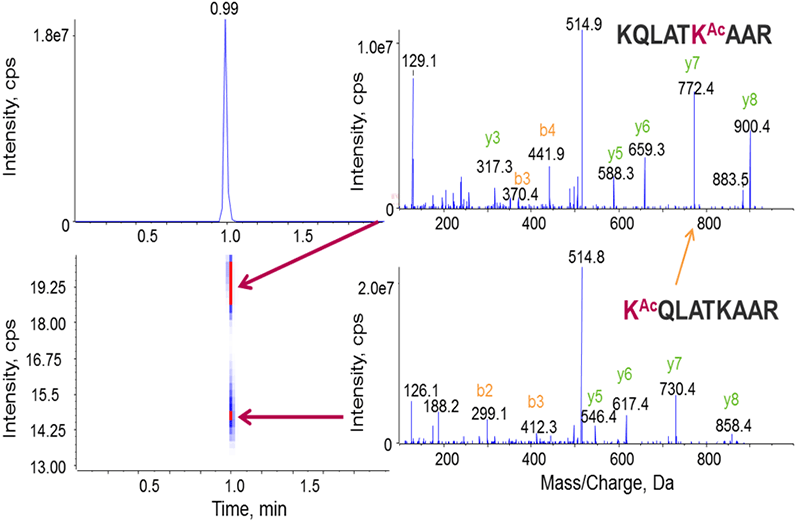 Click to enlarge
Click to enlarge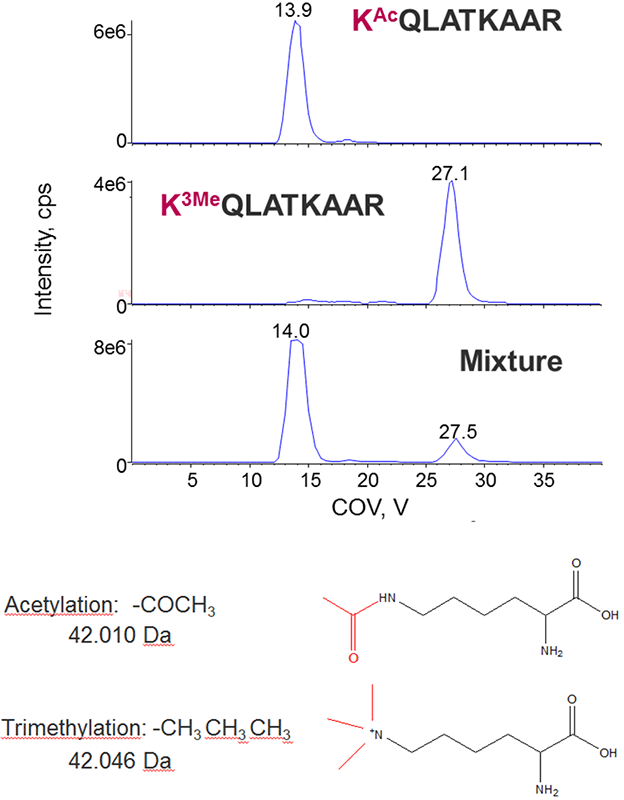 Click to enlarge
Click to enlarge