Discovery to validation: transition from SWATH® Acquisition to targeted MRM analysis for quantitative proteomics pipeline
Using the TripleTOF® 5600+ System and QTRAP® 6500 System
Sahana Mollah, Christie Hunter
SCIEX, USA
Introduction
In their efforts to accurately quantify and verify putative biomarkers or other proteins of interest, researchers often move from discovery techniques, such as shotgun proteomics, to targeted assays in order to obtain accurate quantitation of analytes in complex mixtures. MRM (Multiple Reaction Monitoring) is the gold standard for performing this type of targeted analysis, providing high sensitivity and selectivity. However, there is a limit to the number of MRMs that can be monitored in a single injection. Data independent acquisition is rapidly growing as a key technique for more comprehensive quantitation.
In their efforts to accurately quantify and verify putative biomarkers or other proteins of interest, researchers often move from discovery techniques, such as shotgun proteomics, to targeted assays in order to obtain accurate quantitation of analytes in complex mixtures. MRM (Multiple Reaction Monitoring) is the gold standard for performing this type of targeted analysis, providing high sensitivity and selectivity. However, there is a limit to the number of MRMs that can be monitored in a single injection. Data independent acquisition is rapidly growing as a key technique for more comprehensive quantitation.
In this study, the workflow of performing SWATH Acquisition for comprehensive quantitation followed by MRM analysis on a subset of targets is demonstrated and the results quality of the two techniques compared.
Key aspects of transitioning from SWATH® Acquisition to MRM Analysis
- SWATH Acquisition on the TripleTOF Systems
- Full scan MS/MS of all detectable ions is acquired, enabling post-acquisition interrogation of data
- No upfront assay development is required
- Comprehensive high quality quantification with confirmation of identity on every detectable protein and peptide
- MRM analysis on a QTRAP System
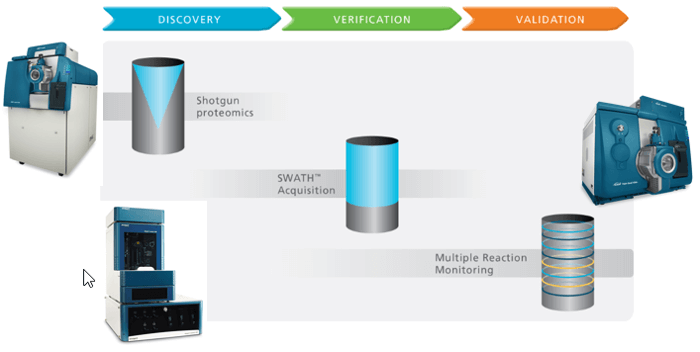
- LINAC® Collision Cell technology in both TripleTOF and QTRAP Systems means that SWATH Acquisition data can be directly transitioned into an MRM assay on the QTRAP 6500 System providing seamless pipeline (Figure 1)
- Higher sensitivity and specificity MRM assays on a subset of proteins and peptides for validation
- Use additional specificity and sensitivity to accelerate assays using faster gradients or higher flow rates2,3
Figure 1. Comprehensive quantitation using SWATH Acquisition followed by high sensitivity MRM Analysis. SWATH Acquisition on a TripleTOF® 5600+ System provides a powerful new approach for discovery and verification experiments. Transition to MRM analysis for high throughput validation on QTRAP® Systems is straightforward.
Experimental
Sample preparation: A tryptic digest of E.coli sample was analyzed with a 1 µg load on column.
Chromatography: The samples were analyzed using the NanoLC™ 425 System in Trap-Elute mode. A elution gradient of 5-35% acetonitrile (0.1% formic acid) in 60 mins was used on a nanoflow column (75 µm x 15 cm ChromXP C18-CL, 3 µm, 300 Å).
Mass spectrometry: SWATH Acquisition was performed using a TripleTOF 5600+ System interfaced to the NanoSpray® Source. The acquisition strategies used was 25Da Q1 windows with 100 msec accumulation time across a mass range of 400-1000 m/z. A set of protein/peptides of interest across an abundance range obtained from SWATH Acquisition data were then chosen for further MRM analysis. Protein/peptide data were loaded into Skyline for MRM assay development on the QTRAP 6500 System. For low intensity proteins with only a single peptide quantified, in silico MRM transitions of other peptides from the same protein were added to the assay, and targeted using MIDAS™ Workflow for sequence confirmation. Replicates were run for both acquisition strategies.
Data processing: SWATH Acquisition data was processed using the PeakView® Software and the SWATH Acquisition MicroApp. MRM data analysis was performed using MultiQuant™ Software and data were compared to SWATH Acquisition data using Excel tools.
Figure 2: Quantitative analysis of SWATH Acquisition replicates. (Top) Fragment XICs with peak areas across ~3.5 orders dynamic range were quantified. (Middle) High reproducibility was achieved on very high numbers of peptides and transitions across the intensity range of peak areas. High intensity peaks showed median CVs as low 2%, with many low intensity peaks still showing good reproducibility of 15%. (Bottom) Cumulative %CV curves show that ~90% of the transition level data has CVs better than 20% and the peptide and protein level data is even better.
High reproducibility SWATH Acquisition
SWATH Acquisition provides an excellent technique to broadly interrogate many proteins and peptides in a sample with very good quantitative quality (Figure 2). Because it is a data independent acquisition strategy, all data are collected using a single acquisition method, with no upfront method development required. Data are then interrogated post-acquisition to select the best peptides and transition for quantitation. High quality data was obtained with SWATH Acquisition across five replicate injections (Figure 2). Across the whole dataset, ~90% of peptide transitions had CVs less than 20%. When assessed relative to the transition peak areas, the highest peak intensities provided the best reproducibility as expected, but even the lowest intensity peaks showed excellent reproducibility.
Figure 3. Skyline for MRM Assay development. A subset of proteins from the SWATH Acquisition data were loaded into Skyline software for MRM assay development. Previous MS/MS results were used as a reference, then fragment ion and collision energy optimization was performed. On the right is the MS/MS spectrum of the highlighted E.coli peptide along with the MRM transitions for the final assay.
Transition subset of proteins to MRM analysis
Once a subset of proteins and peptides are found to be of interest and need to be investigated further, they can easily be transitioned to an MRM assay for a higher throughput, higher sensitivity assay. In this case, Skyline was used to develop the MRM assay on a subset of the lower abundance peptides (Figure 3). As both the TripleTOF® and QTRAP® Systems utilize similar collision cell technology, the MS/MS data obtained from the SWATH® Acquisition can be mined to directly inform the building of robust MRM assays.
Figure 4. %CV Comparison between SWATH® Acquisition data and MRM Data for E. coli analyses. (Top) The reproducibility of peptides analyzed by both techniques was compared for 1 µg on column load. The focus in the MRM dataset was on quantifying lower abundance peptides that were detected by the SWATH Acquisition study, and in this regime the MRM data provides a nice improvement in data quality, namely reproducibility. Orange arrow highlights peptide example expanded in Figure 5. (Bottom) Thus, for low abundance proteins the %CV was observed to be better in the MRM data. Plot was created using an approximate intensity correction factor on the MRM data to account for the higher sensitivity.
Comparing results quality
As the same peptides were analyzed by the two techniques, it provides the opportunity to compare the results quality. Most of the SWATH Acquisition data has very low CVs for the replicate analysis, as seen in Figure 2. If the SWATH Acquisition data is analyzed considering peak intensity (Figure 4, bottom), it is observed that the reproducibility is high across the abundance range of the proteins analyzed, but shows a small decay as the peak intensity decreases. The same analysis was done for the MRM data (after performing an intensity correction on the data to account for sensitivity difference between the instruments4) to overlay against the SWATH Acquisition data. One can see that the SWATH Acquisition data is very similar to the MRM data quality across the abundance range, except for the lowest abundance peaks where the added MRM specificity and sensitivity is advantageous.
The CVs obtained for individual peptides that were analyzed in both techniques were compared and plotted in Figure 4 (top). Again, many of the peptides showed very similar CVs between the two techniques showing the high quantitative quality obtained by both techniques. However, some peptides showed improved quantitation using MRM, such as the peptide example highlighted with the orange arrow. The data for this peptide is displayed in Figure 5 (bottom) and shows that for the low abundance transitions, moving to the targeted MRM assay provided improved data quality. This is expected as the QTRAP 6500 System is more sensitive than the TripleTOF® 5600+ system, and is operating with a tighter Q1 window for added specificity (improved S/N, see Figure 5).
Figure 5. Signal comparison of low abundance peptides. (Top) On the left are the XICs of the MS/MS fragments of the peptide GDGTTGEDITSNVR from E. coli DNA Ligase protein obtained from SWATH Acquisition data. It is low level but still provides a %CV close to 10% on the summed fragment ion transitions. In contrast, when the same amount of sample is loaded on the QTRAP 6500 System for analysis, the MRM signal displays higher specificity, with a CV of 3% as shown in the right panel. (Bottom) Again the SWATH Acquisition XICs (left) are compared to those obtained from the MRM analysis (right). This peptide is VGLETLLGVLNASSR from E. coli High Frequency Lysogenization protein and is highlighted with an orange arrow on Figure 4. Low level proteins with some interference in the SWATH Acquisition data can be improved with the higher sensitivity and specificity of MRM analysis.
Scale and completeness of data
Another important aspect of quantitation is the completeness of measurements across all samples - a key advantage of targeted quantitation. It is highly desirable to have a good measurement for every peptide and protein targeted in every sample analyzed. Figure 6 provides a detailed analysis of the data completeness observed across the 5 replicates for the SWATH Acquisition experiment. The blue text show that ~19,500 fragment ions were extracted and 78% of targeted fragments had good peak areas and provided a peak group score of 1% FDR (Figure 6, top). Once fragment ions are summed to the peptide and protein level, the data completeness approaches 95% or higher. The MRM assay was developed to focus on the lower abundance peptides, resulting in a smaller number of peptides targeted (175 peptides), but with a higher success rate of ~91% at the transition level in this case.
Figure 6. Data completeness. These data tables provide a summary of the experiments to highlight the differences between scale and efficiency of the two techniques. In the SWATH Acquisition experiment (top), over 4500 peptides and 19000 transitions were analyzed across 5 sample replicates. By looking at each row in the feature table and counting how many peak areas were obtained, the data completeness was measured at 78%. In the MRM experiment (bottom), a much smaller number of peptides were targeted (175), but higher data completeness was observed at 91%.
Conclusions
High quantitative reproducibility on very large numbers of peptides and proteins can be obtained using SWATH Acquisition on the TripleTOF® 5600+ System. By targeting a smaller subset of peptides using MRM analysis on a QTRAP® 6500 System, a further improvement in quantitative quality can be achieved, especially on for lower abundance proteins. This work demonstrates the ability to perform initial quantification (verification) on large numbers of proteins using SWATH Acquisition, then easily transition to an improved targeted MRM assay for accelerated quantification.
References
- Improved Data Quality Using Variable Q1 Window Widths in SWATH® Acquisition. SCIEX technical note RUO-MKT-02-2879-B.
- Exploring the Sensitivity Differences for Peptide Quantification in the Low Flow Rate Regime - nanoLC™ 400 System for High Performance Nanoflow and Microflow LC. SCIEX technical note RUO-MKT-02-3252-A.
- Achieve Low Flow Sensitivities with Micro Flow Chromatography on the QTRAP® 6500 System for Targeted Quantitative Proteomics. SCIEX technical note RUO-MKT-02-3296-A.
- In order to overlay the %CV curves for both instruments, a correction factor was applied to adjust the X-axis for the signal intensity difference between the two instruments (~10x).
 Click to enlarge
Click to enlarge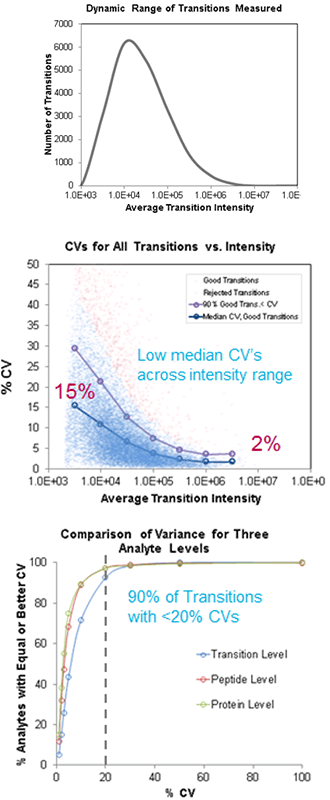 Click to enlarge
Click to enlarge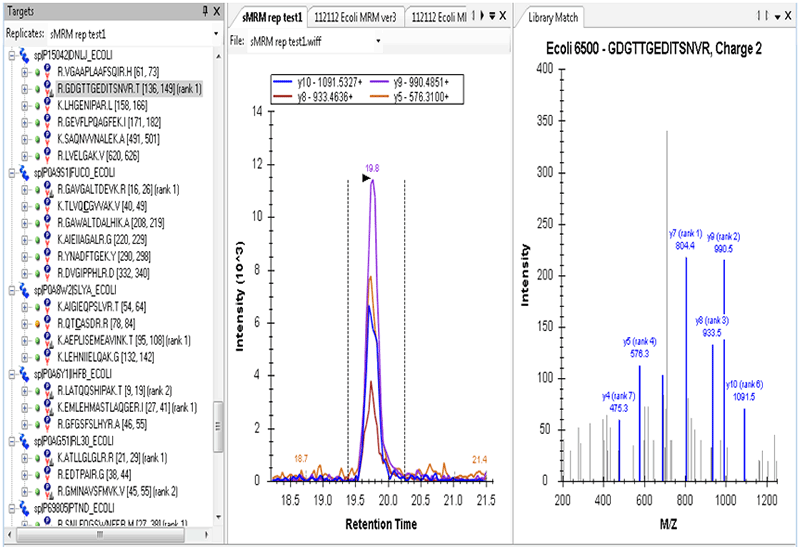 Click to enlarge
Click to enlarge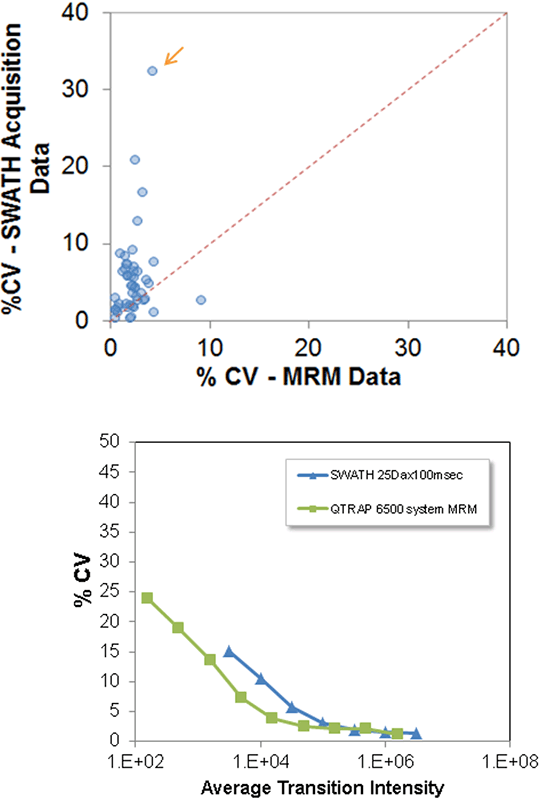 Click to enlarge
Click to enlarge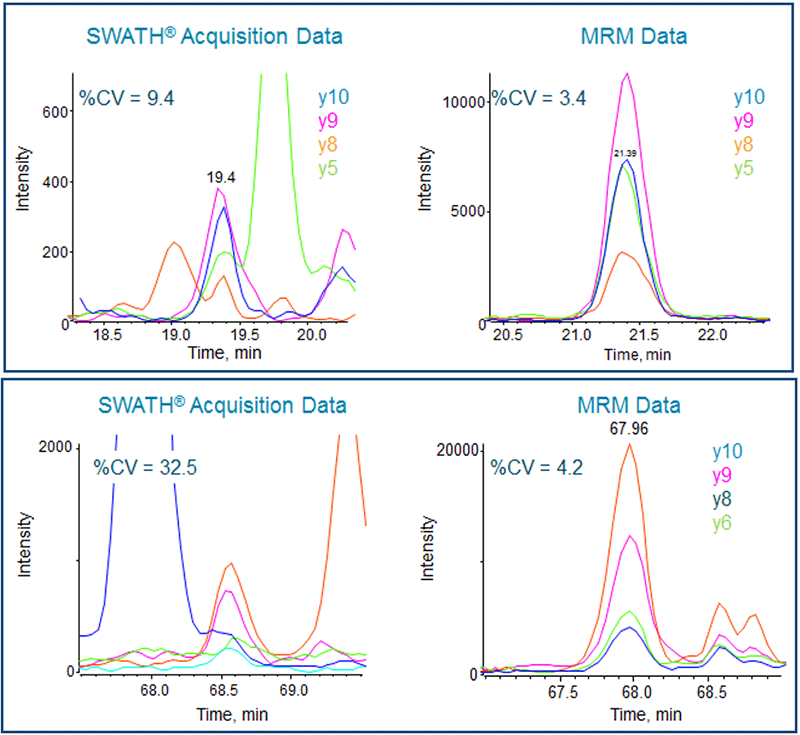 Click to enlarge
Click to enlarge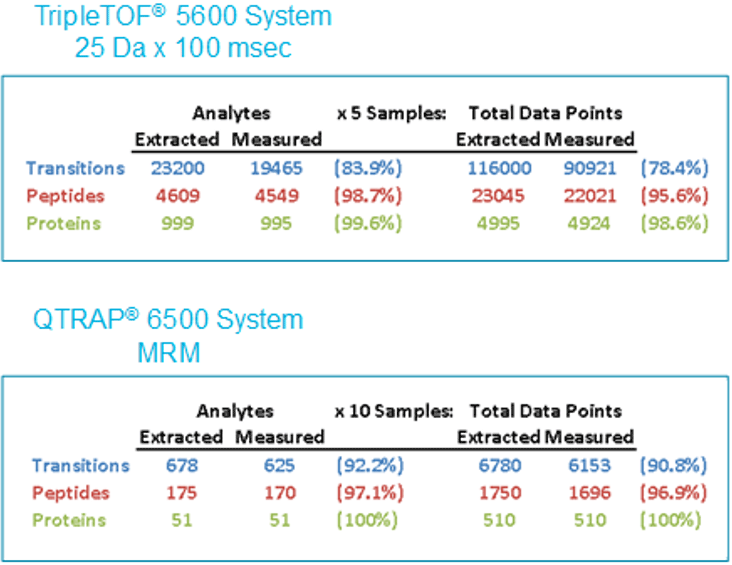 Click to enlarge
Click to enlarge