Spectral library generation for SWATH® Acquisition in less than 20 hours
Using the OneOmics™ Suite and the TripleTOF® 6600+ LC-MS/MS System
Nick Morrice1, and Christie Hunter2
1SCIEX UK, 2SCIEX, USA
Abstract
Using fast microflow LC on the TripleTOF 6600+ System combined with cloud processing with OneOmics Suite in SCIEX Cloud Platform, protein identification experiments or generation of ion libraries can be rapidly generated. In under 20 hours, LC-MS data was acquired on 40 proteome fractions, then uploaded to the cloud and searched using the ProteinPilot App to generate very good protein identification results.

Introduction
The generation of spectral libraries is often thought of as a time consuming procedure, but in reality it can be performed in less than 1 day. It has been shown previously that utilizing rapid gradients and microflow chromatography can provide very good protein and peptide identification numbers.1 It is also a common understanding in the field of proteomics that many more protein identifications can be generated by first performing high pH reversed phase fractionation of complex digested samples, running data dependent analysis of each fraction and then searching all the LC-MS/MS data together to generate a very large ion library.2 The database searches for protein identifications can often be the rate limiting factor even when using powerful desktop computers as the datasets get very large.
Here, the combination of in-depth sample fractionation with microflow LC on a TripleTOF 6600+ System and cloud computing was explored for the rapid generation of bespoke libraries. Two sample types were tested, plasma and a tissue lysate.
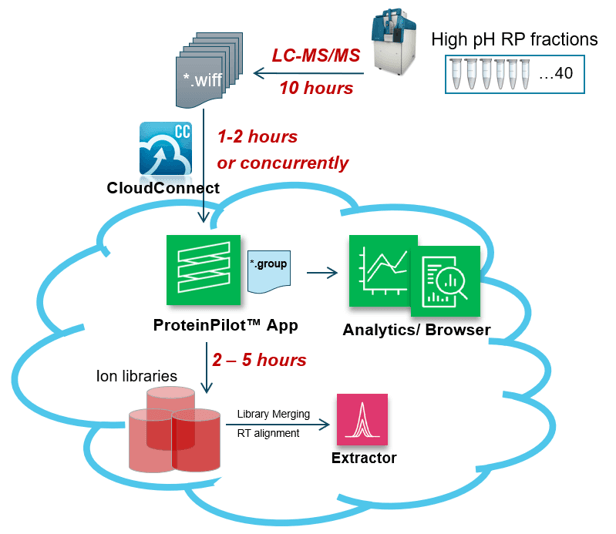
Figure 1. Workflow for fast ion library generation on proteomic samples. After using high pH fractionation to generate 40 fractions, they were rapidly analyzed by LC-MS/MS using microflow LC. Data were uploaded to the cloud for processing in order to generate a large ion library for SWATH® Acquisition data processing.
Key features of the fast library generation strategy
- Full workflow from sample to ion library took less than 24 hours (Figure 1)
- Forty high pH fractions were used for sub-fractionation to deeply interrogate the samples in order to generate large ion libraries
- Fast microflow LC gradients (10 mins) were used to then rapidly analyze the 40 fractions
- The TripleTOF 6600+ System was used and operated at 50 Hz to generate as much high-quality, high-resolution MS/MS data as possible
- Data were uploaded to SCIEX Cloud Platform and searched with the ProteinPilot™ App to identify a large number of proteins from the fractions and generate an ion library for SWATH® Acquisition processing
 Click to enlarge
Click to enlarge Click to enlarge
Click to enlarge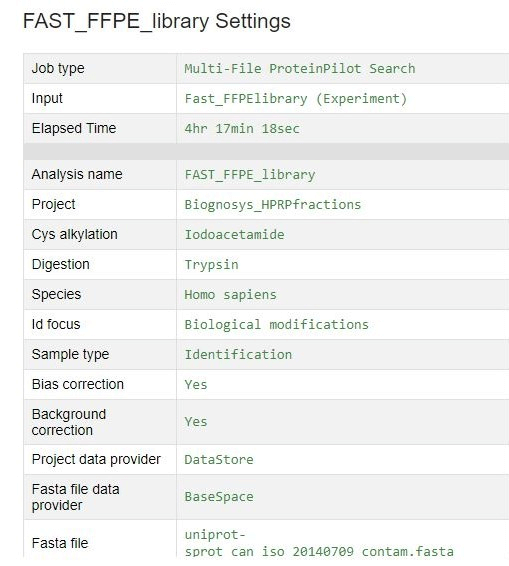 Click to enlarge
Click to enlarge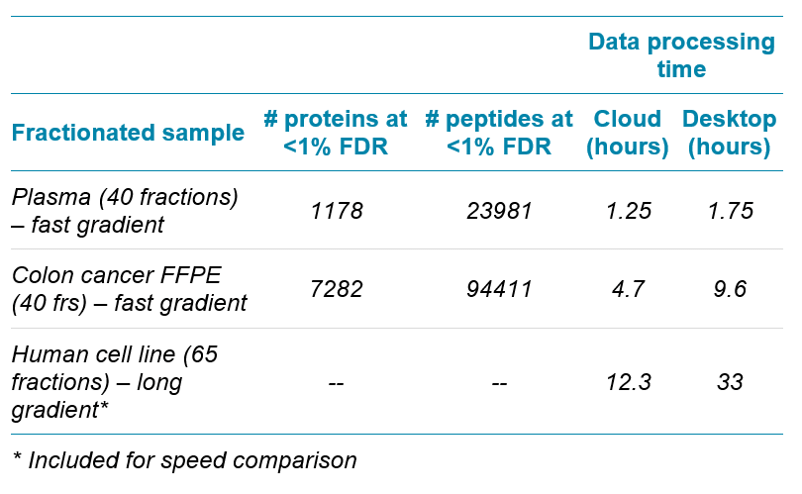 Click to enlarge
Click to enlarge